Transcriptional pausing controls a rapid antiviral innate immune response in Drosophila
- PMID: 23084920
- PMCID: PMC3479682
- DOI: 10.1016/j.chom.2012.08.011
Transcriptional pausing controls a rapid antiviral innate immune response in Drosophila
Abstract
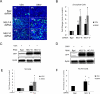
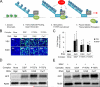
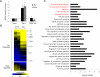
Innate immune responses are characterized by precise gene expression whereby gene subsets are temporally induced to limit infection, although the mechanisms involved are incompletely understood. We show that antiviral immunity in Drosophila requires the transcriptional pausing pathway, including negative elongation factor (NELF) that pauses RNA polymerase II (Pol II) and positive elongation factor b (P-TEFb), which releases paused Pol II to produce full-length transcripts. We identify a set of genes that is rapidly transcribed upon arbovirus infection, including components of antiviral pathways (RNA silencing, autophagy, JAK/STAT, Toll, and Imd) and various Toll receptors. Many of these genes require P-TEFb for expression and exhibit pausing-associated chromatin features. Furthermore, transcriptional pausing is critical for antiviral immunity in insects because NELF and P-TEFb are required to restrict viral replication in adult flies and vector mosquito cells. Thus, transcriptional pausing primes virally induced genes to facilitate rapid gene induction and robust antiviral responses.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
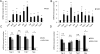


Similar articles
-
PTEN modulates gene transcription by redistributing genome-wide RNA polymerase II occupancy.Hum Mol Genet. 2019 Sep 1;28(17):2826-2834. doi: 10.1093/hmg/ddz112. Hum Mol Genet. 2019. PMID: 31127935 Free PMC article.
-
Nup98 promotes antiviral gene expression to restrict RNA viral infection in Drosophila.Proc Natl Acad Sci U S A. 2014 Sep 16;111(37):E3890-9. doi: 10.1073/pnas.1410087111. Epub 2014 Sep 2. Proc Natl Acad Sci U S A. 2014. PMID: 25197089 Free PMC article.
-
[Antiviral immunity in drosophila].J Soc Biol. 2007;201(4):359-65. doi: 10.1051/jbio:2007906. Epub 2008 Mar 5. J Soc Biol. 2007. PMID: 18533096 French.
-
Genetic determinants of antiviral immunity in dipteran insects - Compiling the experimental evidence.Dev Comp Immunol. 2021 Jun;119:104010. doi: 10.1016/j.dci.2021.104010. Epub 2021 Jan 19. Dev Comp Immunol. 2021. PMID: 33476667 Review.
-
Mosquito RNAi is the major innate immune pathway controlling arbovirus infection and transmission.Future Microbiol. 2011 Mar;6(3):265-77. doi: 10.2217/fmb.11.11. Future Microbiol. 2011. PMID: 21449839 Free PMC article. Review.
Cited by
-
Viruses and antiviral immunity in Drosophila.Dev Comp Immunol. 2014 Jan;42(1):67-84. doi: 10.1016/j.dci.2013.05.002. Epub 2013 May 13. Dev Comp Immunol. 2014. PMID: 23680639 Free PMC article. Review.
-
Induced antiviral innate immunity in Drosophila.Curr Opin Microbiol. 2014 Aug;20:62-8. doi: 10.1016/j.mib.2014.05.006. Epub 2014 Jun 5. Curr Opin Microbiol. 2014. PMID: 24907422 Free PMC article. Review.
-
Innate and intrinsic antiviral immunity in Drosophila.Cell Mol Life Sci. 2017 Jun;74(11):2039-2054. doi: 10.1007/s00018-017-2453-9. Epub 2017 Jan 19. Cell Mol Life Sci. 2017. PMID: 28102430 Free PMC article. Review.
-
Investigating the Evolution of Drosophila STING-Dependent Antiviral Innate Immunity by Multispecies Comparison of 2'3'-cGAMP Responses.Mol Biol Evol. 2024 Mar 1;41(3):msae032. doi: 10.1093/molbev/msae032. Mol Biol Evol. 2024. PMID: 38377349 Free PMC article.
-
Hexamethylene bisacetamide impairs NK cell-mediated clearance of acute T lymphoblastic leukemia cells and HIV-1-infected T cells that exit viral latency.Sci Rep. 2019 Mar 13;9(1):4373. doi: 10.1038/s41598-019-40760-x. Sci Rep. 2019. PMID: 30867508 Free PMC article.
References
-
- Adelman K, Kennedy MA, Nechaev S, Gilchrist DA, Muse GW, Chinenov Y, Rogatsky I. Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18207–18212. - PMC - PubMed
-
- Adelman K, Rogatsky I. RNA polymerase II stalling mediates cytokine gene expression. Cell Cycle. 2010;9:630–631. - PubMed
-
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

