Short-range exosomal transfer of viral RNA from infected cells to plasmacytoid dendritic cells triggers innate immunity
- PMID: 23084922
- PMCID: PMC3479672
- DOI: 10.1016/j.chom.2012.08.010
Short-range exosomal transfer of viral RNA from infected cells to plasmacytoid dendritic cells triggers innate immunity
Abstract
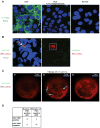
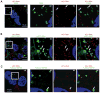
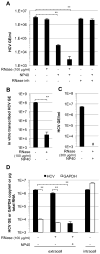
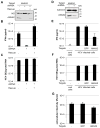
Viral nucleic acids often trigger an innate immune response in infected cells. Many viruses, including hepatitis C virus (HCV), have evolved mechanisms to evade intracellular recognition. Nevertheless, HCV-permissive cells can trigger a viral RNA-, TLR7-, and cell-contact-dependent compensatory interferon response in nonpermissive plasmacytoid dendritic cells (pDCs). Here we report that these events are mediated by transfer of HCV-RNA-containing exosomes from infected cells to pDCs. The exosomal viral RNA transfer is dependent on the endosomal sorting complex required for transport (ESCRT) machinery and on Annexin A2, an RNA-binding protein involved in membrane vesicle trafficking, and is suppressed by exosome release inhibitors. Further, purified concentrated HCV-RNA-containing exosomes are sufficient to activate pDCs. Thus, vesicular sequestration and exosomal export of viral RNA may serve both as a viral strategy to evade pathogen sensing within infected cells and as a host strategy to induce an unopposed innate response in replication-nonpermissive bystander cells.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures



Similar articles
-
The Dual Role of Exosomes in Hepatitis A and C Virus Transmission and Viral Immune Activation.Viruses. 2015 Dec 17;7(12):6707-15. doi: 10.3390/v7122967. Viruses. 2015. PMID: 26694453 Free PMC article. Review.
-
Plasmacytoid dendritic cells sense hepatitis C virus-infected cells, produce interferon, and inhibit infection.Proc Natl Acad Sci U S A. 2010 Apr 20;107(16):7431-6. doi: 10.1073/pnas.1002301107. Epub 2010 Mar 15. Proc Natl Acad Sci U S A. 2010. PMID: 20231459 Free PMC article.
-
Hepatitis C virus pathogen associated molecular pattern (PAMP) triggers production of lambda-interferons by human plasmacytoid dendritic cells.PLoS Pathog. 2013;9(4):e1003316. doi: 10.1371/journal.ppat.1003316. Epub 2013 Apr 18. PLoS Pathog. 2013. PMID: 23637605 Free PMC article.
-
Sensing of latent EBV infection through exosomal transfer of 5'pppRNA.Proc Natl Acad Sci U S A. 2016 Feb 2;113(5):E587-96. doi: 10.1073/pnas.1518130113. Epub 2016 Jan 14. Proc Natl Acad Sci U S A. 2016. PMID: 26768848 Free PMC article.
-
Extracellular Vesicles Deliver Host and Virus RNA and Regulate Innate Immune Response.Int J Mol Sci. 2017 Mar 20;18(3):666. doi: 10.3390/ijms18030666. Int J Mol Sci. 2017. PMID: 28335522 Free PMC article. Review.
Cited by
-
Proteomics of Human Dendritic Cell Subsets Reveals Subset-Specific Surface Markers and Differential Inflammasome Function.Cell Rep. 2016 Sep 13;16(11):2953-2966. doi: 10.1016/j.celrep.2016.08.023. Cell Rep. 2016. PMID: 27626665 Free PMC article.
-
CD169+ macrophages orchestrate plasmacytoid dendritic cell arrest and retention for optimal priming in the bone marrow of malaria-infected mice.Elife. 2022 Oct 24;11:e78873. doi: 10.7554/eLife.78873. Elife. 2022. PMID: 36278864 Free PMC article.
-
Tracking Plasmacytoid Dendritic Cell Response to Physical Contact with Infected Cells.Methods Mol Biol. 2023;2618:289-315. doi: 10.1007/978-1-0716-2938-3_21. Methods Mol Biol. 2023. PMID: 36905525
-
The immunological function of extracellular vesicles in hepatitis B virus-infected hepatocytes.PLoS One. 2018 Dec 31;13(12):e0205886. doi: 10.1371/journal.pone.0205886. eCollection 2018. PLoS One. 2018. PMID: 30596665 Free PMC article.
-
Multifunctional role of exosomes in viral diseases: From transmission to diagnosis and therapy.Cell Signal. 2022 Jun;94:110325. doi: 10.1016/j.cellsig.2022.110325. Epub 2022 Mar 31. Cell Signal. 2022. PMID: 35367363 Free PMC article. Review.
References
-
- Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, Rak J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10:619–624. - PubMed
-
- Alter H, Seeff LB. Recovery, persistence, and sequelae in hepatitis C virus infection: a perspective on long-term outcome. Semin Liver Dis. 2000;20:17–35. - PubMed
-
- Asselin-Paturel C, Brizard G, Pin JJ, Briere F, Trinchieri G. Mouse strain differences in plasmacytoid dendritic cell frequency and function revealed by a novel monoclonal antibody. J Immunol. 2003;171:6466–6477. - PubMed
-
- Aukrust I, Hollas H, Strand E, Evensen L, Trave G, Flatmark T, Vedeler A. The mRNA-binding site of annexin A2 resides in helices C-D of its domain IV. J Mol Biol. 2007;368:1367–1378. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

