Generalized disruption of inherited genomic imprints leads to wide-ranging placental defects and dysregulated fetal growth
- PMID: 23085235
- PMCID: PMC3508140
- DOI: 10.1016/j.ydbio.2012.10.010
Generalized disruption of inherited genomic imprints leads to wide-ranging placental defects and dysregulated fetal growth
Abstract
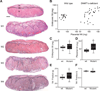
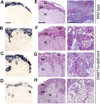
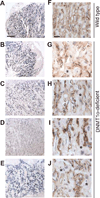
Monoallelic expression of imprinted genes, including ones solely expressed in the placenta, is essential for normal placental development and fetal growth. To better understand the role of placental imprinting in placental development and fetal growth, we examined conceptuses developing in the absence of maternally derived DNA (cytosine-5-)-methyltransferase 1o (DNMT1o). Absence of DNMT1o results in the partial loss of methylation at imprinted differentially methylated domain (DMD) sequences in the embryo and the placenta. Mid-gestation E9.5 DNMT1o-deficient placentas exhibited structural abnormalities of all tissue layers. At E17.5, all examined placentas had aberrant placental morphology, most notably in the spongiotrophoblast and labyrinth layers. Abnormalities included an expanded volume fraction of spongiotrophoblast tissue with extension of the spongiotrophoblast layer into the labyrinth. Many mutant placentas also demonstrated migration abnormalities of glycogen cells. Additionally, the volume fraction of the labyrinth was reduced, as was the surface area for maternal fetal gas exchange. Despite these placental morphologic abnormalities, approximately one-half of DNMT1o-deficient fetuses survived to late gestation (E17.5). Furthermore, DNMT1o-deficient placentas supported a broad range of fetal growth. The ability of some DNMT1o-deficient and morphologically abnormal placentas to support fetal growth in excess of wild type demonstrates the importance of differential methylation of DMDs and proper imprinting of discrete gene clusters to placental morphogenesis and fetal growth.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures




Similar articles
-
Loss of DNMT1o disrupts imprinted X chromosome inactivation and accentuates placental defects in females.PLoS Genet. 2013 Nov;9(11):e1003873. doi: 10.1371/journal.pgen.1003873. Epub 2013 Nov 21. PLoS Genet. 2013. PMID: 24278026 Free PMC article.
-
Loss of inherited genomic imprints in mice leads to severe disruption in placental lipid metabolism.Placenta. 2015 Apr;36(4):389-96. doi: 10.1016/j.placenta.2015.01.012. Epub 2015 Jan 29. Placenta. 2015. PMID: 25662615 Free PMC article.
-
Partial Loss of Genomic Imprinting Reveals Important Roles for Kcnq1 and Peg10 Imprinted Domains in Placental Development.PLoS One. 2015 Aug 4;10(8):e0135202. doi: 10.1371/journal.pone.0135202. eCollection 2015. PLoS One. 2015. PMID: 26241757 Free PMC article.
-
Genomic imprinting, action, and interaction of maternal and fetal genomes.Proc Natl Acad Sci U S A. 2015 Jun 2;112(22):6834-40. doi: 10.1073/pnas.1411253111. Epub 2014 Nov 17. Proc Natl Acad Sci U S A. 2015. PMID: 25404322 Free PMC article. Review.
-
Epigenetic regulation and related diseases during placental development.Yi Chuan. 2017 Apr 20;39(4):263-275. doi: 10.16288/j.yczz.16-380. Yi Chuan. 2017. PMID: 28420606 Review.
Cited by
-
DNA methylation across the tree of life, from micro to macro-organism.Bioengineered. 2022 Jan;13(1):1666-1685. doi: 10.1080/21655979.2021.2014387. Bioengineered. 2022. PMID: 34986742 Free PMC article.
-
Loss of DNMT1o disrupts imprinted X chromosome inactivation and accentuates placental defects in females.PLoS Genet. 2013 Nov;9(11):e1003873. doi: 10.1371/journal.pgen.1003873. Epub 2013 Nov 21. PLoS Genet. 2013. PMID: 24278026 Free PMC article.
-
Loss-of-function mutation of NSD2 is associated with abnormal placentation accompanied by fetal growth retardation in mice.PLoS One. 2025 Jul 21;20(7):e0328243. doi: 10.1371/journal.pone.0328243. eCollection 2025. PLoS One. 2025. PMID: 40690504 Free PMC article.
-
Transient DNMT1 suppression reveals hidden heritable marks in the genome.Nucleic Acids Res. 2015 Feb 18;43(3):1485-97. doi: 10.1093/nar/gku1386. Epub 2015 Jan 10. Nucleic Acids Res. 2015. PMID: 25578964 Free PMC article.
-
Loss of inherited genomic imprints in mice leads to severe disruption in placental lipid metabolism.Placenta. 2015 Apr;36(4):389-96. doi: 10.1016/j.placenta.2015.01.012. Epub 2015 Jan 29. Placenta. 2015. PMID: 25662615 Free PMC article.
References
-
- Baddeley AJ, Gundersen HJ, Cruz-Orive LM. Estimation of surface area from vertical sections. J. Microsc. 1986;142:259–276. - PubMed
-
- Barak Y, Nelson MC, Ong ES, Jones YZ, Ruiz-Lozano P, Chien KR, Koder A, Evans RM. PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol. Cell. 1999;4:585–595. - PubMed
-
- Bartolomei MS, Tilghman SM. Genomic imprinting in mammals. Annu. Rev. Genet. 1997;31:493–525. - PubMed
-
- Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

