Spatiotemporal regulation of an Hcn4 enhancer defines a role for Mef2c and HDACs in cardiac electrical patterning
- PMID: 23085412
- PMCID: PMC3510001
- DOI: 10.1016/j.ydbio.2012.10.017
Spatiotemporal regulation of an Hcn4 enhancer defines a role for Mef2c and HDACs in cardiac electrical patterning
Abstract
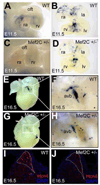
Regional differences in cardiomyocyte automaticity permit the sinoatrial node (SAN) to function as the leading cardiac pacemaker and the atrioventricular (AV) junction as a subsidiary pacemaker. The regulatory mechanisms controlling the distribution of automaticity within the heart are not understood. To understand regional variation in cardiac automaticity, we carried out an in vivo analysis of cis-regulatory elements that control expression of the hyperpolarization-activated cyclic-nucleotide gated ion channel 4 (Hcn4). Using transgenic mice, we found that spatial and temporal patterning of Hcn4 expression in the AV conduction system required cis-regulatory elements with multiple conserved fragments. One highly conserved region, which contained a myocyte enhancer factor 2C (Mef2C) binding site previously described in vitro, induced reporter expression specifically in the embryonic non-chamber myocardium and the postnatal AV bundle in a Mef2c-dependent manner in vivo. Inhibition of histone deacetylase (HDAC) activity in cultured transgenic embryos showed expansion of reporter activity to working myocardium. In adult animals, hypertrophy induced by transverse aortic constriction, which causes translocation of HDACs out of the nucleus, resulted in ectopic activation of the Hcn4 enhancer in working myocardium, recapitulating pathological electrical remodeling. These findings reveal mechanisms that control the distribution of automaticity among cardiomyocytes during development and in response to stress.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures








References
-
- Aanhaanen WT, Mommersteeg MT, Norden J, Wakker V, de Gier-de Vries C, Anderson RH, Kispert A, Moorman AF, Christoffels VM. Developmental origin, growth, and three-dimensional architecture of the atrioventricular conduction axis of the mouse heart. Circ Res. 2010;107:728–736. - PubMed
-
- Bakker ML, Boukens BJ, Mommersteeg MT, Brons JF, Wakker V, Moorman AF, Christoffels VM. Transcription factor Tbx3 is required for the specification of the atrioventricular conduction system. Circ Res. 2008a;102:1340–1349. - PubMed
-
- Bakker ML, Boukens BJ, Mommersteeg MTM, Brons JF, Wakker V, Moorman AFM, Christoffels VM. Transcription factor Tbx3 is required for the specification of the atrioventricular conduction system. Circulation Research. 2008b;102:1340–1349. - PubMed
-
- Bakker ML, Moorman AF, Christoffels VM. The atrioventricular node: origin, development, and genetic program. Trends Cardiovasc Med. 2011;20:164–171. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

