Multimodal microtubule binding by the Ndc80 kinetochore complex
- PMID: 23085714
- PMCID: PMC3492541
- DOI: 10.1038/nsmb.2411
Multimodal microtubule binding by the Ndc80 kinetochore complex
Abstract
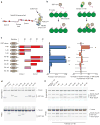
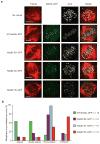
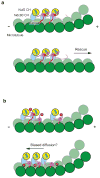
The Ndc80 complex is a key site of kinetochore-microtubule attachment during cell division. The human complex engages microtubules with a globular 'head' formed by tandem calponin-homology domains and an 80-amino-acid unstructured 'tail' that contains sites of phosphoregulation by the Aurora B kinase. Using biochemical, cell biological and electron microscopy analyses, we dissected the roles of the tail in binding of microtubules and mediation of cooperative interactions between Ndc80 complexes. Two segments of the tail that contain Aurora B phosphorylation sites become ordered at interfaces; one with tubulin and the second with an adjacent Ndc80 head on the microtubule surface, forming interactions that are disrupted by phosphorylation. We propose a model in which Ndc80's interaction with either growing or shrinking microtubule ends can be tuned by the phosphorylation state of its tail.
Figures



References
-
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–97. - PubMed
-
- McCleland ML, et al. The vertebrate Ndc80 complex contains Spc24 and Spc25 homologs, which are required to establish and maintain kinetochore–microtubule attachment. Curr Biol. 2004;14:131–7. - PubMed
-
- DeLuca JG, et al. Kinetochore microtubule dynamics and attachment stability are regulated by HEC1. Cell. 2006;127:969–82. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

