The Arabidopsis thylakoid ADP/ATP carrier TAAC has an additional role in supplying plastidic phosphoadenosine 5'-phosphosulfate to the cytosol
- PMID: 23085732
- PMCID: PMC3517245
- DOI: 10.1105/tpc.112.101964
The Arabidopsis thylakoid ADP/ATP carrier TAAC has an additional role in supplying plastidic phosphoadenosine 5'-phosphosulfate to the cytosol
Abstract
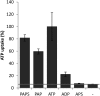
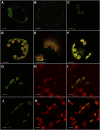
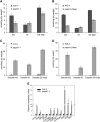
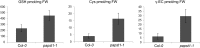
3'-Phosphoadenosine 5'-phosphosulfate (PAPS) is the high-energy sulfate donor for sulfation reactions. Plants produce some PAPS in the cytosol, but it is predominantly produced in plastids. Accordingly, PAPS has to be provided by plastids to serve as a substrate for sulfotransferase reactions in the cytosol and the Golgi apparatus. We present several lines of evidence that the recently described Arabidopsis thaliana thylakoid ADP/ATP carrier TAAC transports PAPS across the plastid envelope and thus fulfills an additional function of high physiological relevance. Transport studies using the recombinant protein revealed that it favors PAPS, 3'-phosphoadenosine 5'-phosphate, and ATP as substrates; thus, we named it PAPST1. The protein could be detected both in the plastid envelope membrane and in thylakoids, and it is present in plastids of autotrophic and heterotrophic tissues. TAAC/PAPST1 belongs to the mitochondrial carrier family in contrast with the known animal PAPS transporters, which are members of the nucleotide-sugar transporter family. The expression of the PAPST1 gene is regulated by the same MYB transcription factors also regulating the biosynthesis of sulfated secondary metabolites, glucosinolates. Molecular and physiological analyses of papst1 mutant plants indicate that PAPST1 is involved in several aspects of sulfur metabolism, including the biosynthesis of thiols, glucosinolates, and phytosulfokines.
Figures





Similar articles
-
PAPST2 Plays Critical Roles in Removing the Stress Signaling Molecule 3'-Phosphoadenosine 5'-Phosphate from the Cytosol and Its Subsequent Degradation in Plastids and Mitochondria.Plant Cell. 2019 Jan;31(1):231-249. doi: 10.1105/tpc.18.00512. Epub 2018 Nov 21. Plant Cell. 2019. PMID: 30464037 Free PMC article.
-
Identification, expression, and functional analyses of a thylakoid ATP/ADP carrier from Arabidopsis.J Biol Chem. 2007 Mar 23;282(12):8848-59. doi: 10.1074/jbc.M609130200. Epub 2007 Jan 29. J Biol Chem. 2007. PMID: 17261580
-
Molecular cloning and identification of 3'-phosphoadenosine 5'-phosphosulfate transporter.J Biol Chem. 2003 Jul 11;278(28):25958-63. doi: 10.1074/jbc.M302439200. Epub 2003 Apr 25. J Biol Chem. 2003. PMID: 12716889
-
Roles of the nucleotide sugar transporters (SLC35 family) in health and disease.Mol Aspects Med. 2013 Apr-Jun;34(2-3):590-600. doi: 10.1016/j.mam.2012.12.004. Mol Aspects Med. 2013. PMID: 23506892 Review.
-
Sulfation and sulfotransferases 5: the importance of 3'-phosphoadenosine 5'-phosphosulfate (PAPS) in the regulation of sulfation.FASEB J. 1997 May;11(6):404-18. doi: 10.1096/fasebj.11.6.9194521. FASEB J. 1997. PMID: 9194521 Review.
Cited by
-
A metabolic, phylogenomic and environmental atlas of diatom plastid transporters from the model species Phaeodactylum.Front Plant Sci. 2022 Sep 22;13:950467. doi: 10.3389/fpls.2022.950467. eCollection 2022. Front Plant Sci. 2022. PMID: 36212359 Free PMC article.
-
PAPST2 Plays Critical Roles in Removing the Stress Signaling Molecule 3'-Phosphoadenosine 5'-Phosphate from the Cytosol and Its Subsequent Degradation in Plastids and Mitochondria.Plant Cell. 2019 Jan;31(1):231-249. doi: 10.1105/tpc.18.00512. Epub 2018 Nov 21. Plant Cell. 2019. PMID: 30464037 Free PMC article.
-
Sulfated plant peptide hormones.J Exp Bot. 2019 Aug 19;70(16):4267-4277. doi: 10.1093/jxb/erz292. J Exp Bot. 2019. PMID: 31231771 Free PMC article. Review.
-
The multi-protein family of sulfotransferases in plants: composition, occurrence, substrate specificity, and functions.Front Plant Sci. 2014 Oct 16;5:556. doi: 10.3389/fpls.2014.00556. eCollection 2014. Front Plant Sci. 2014. PMID: 25360143 Free PMC article. Review.
-
Catalysts for sulfur: understanding the intricacies of enzymes orchestrating plant sulfur anabolism.Planta. 2024 Dec 17;261(1):16. doi: 10.1007/s00425-024-04594-w. Planta. 2024. PMID: 39690279 Review.
References
-
- Andrès C., Agne B., Kessler F. (2010). The TOC complex: Preprotein gateway to the chloroplast. Biochim. Biophys. Acta 1803: 715–723 - PubMed
-
- Benderoth M., Pfalz M., Kroymann J. (2009). Methylthioalkylmalate synthases: Genetics, ecology and evolution. Phytochem. Rev. 8: 255–268
-
- Bedhomme M., Hoffmann M., McCarthy E.A., Gambonnet B., Moran R.G., Rébeillé F., Ravanel S. (2005). Folate metabolism in plants: An Arabidopsis homolog of the mammalian mitochondrial folate transporter mediates folate import into chloroplasts. J. Biol. Chem. 280: 34823–34831 - PubMed
-
- Berger B., Stracke R., Yatusevich R., Weisshaar B., Flügge U.I., Gigolashvili T. (2007). A simplified method for the analysis of transcription factor-promoter interactions that allows high-throughput data generation. Plant J. 50: 911–916 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

