Spatio-temporal cellular dynamics of the Arabidopsis flagellin receptor reveal activation status-dependent endosomal sorting
- PMID: 23085733
- PMCID: PMC3516521
- DOI: 10.1105/tpc.112.100263
Spatio-temporal cellular dynamics of the Arabidopsis flagellin receptor reveal activation status-dependent endosomal sorting
Abstract
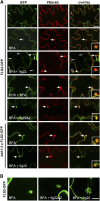
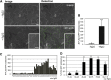
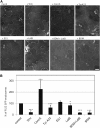
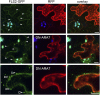
The activity of surface receptors is location specific, dependent upon the dynamic membrane trafficking network and receptor-mediated endocytosis (RME). Therefore, the spatio-temporal dynamics of RME are critical to receptor function. The plasma membrane receptor flagellin sensing2 (FLS2) confers immunity against bacterial infection through perception of flagellin (flg22). Following elicitation, FLS2 is internalized into vesicles. To resolve FLS2 trafficking, we exploited quantitative confocal imaging for colocalization studies and chemical interference. FLS2 localizes to bona fide endosomes via two distinct endocytic trafficking routes depending on its activation status. FLS2 receptors constitutively recycle in a Brefeldin A (BFA)-sensitive manner, while flg22-activated receptors traffic via ARA7/Rab F2b- and ARA6/Rab F1-positive endosomes insensitive to BFA. FLS2 endocytosis required a functional Rab5 GTPase pathway as revealed by dominant-negative ARA7/Rab F2b. Flg22-induced FLS2 endosomal numbers were increased by Concanamycin A treatment but reduced by Wortmannin, indicating that activated FLS2 receptors are targeted to late endosomes. RME inhibitors Tyrphostin A23 and Endosidin 1 altered but did not block induced FLS2 endocytosis. Additional inhibitor studies imply the involvement of the actin-myosin system in FLS2 internalization and trafficking. Altogether, we report a dynamic pattern of subcellular trafficking for FLS2 and reveal a defined framework for ligand-dependent endocytosis of this receptor.
Figures





References
-
- Banbury D.N., Oakley J.D., Sessions R.B., Banting G. (2003). Tyrphostin A23 inhibits internalization of the transferrin receptor by perturbing the interaction between tyrosine motifs and the medium chain subunit of the AP-2 adaptor complex. J. Biol. Chem. 278: 12022–12028 - PubMed
-
- Bandmann V., Homann U. (2012). Clathrin-independent endocytosis contributes to uptake of glucose into BY-2 protoplasts. Plant J. 70: 578–584 - PubMed
-
- Bauer Z., Gómez-Gómez L., Boller T., Felix G. (2001). Sensitivity of different ecotypes and mutants of Arabidopsis thaliana toward the bacterial elicitor flagellin correlates with the presence of receptor-binding sites. J. Biol. Chem. 276: 45669–45676 - PubMed
-
- Bolte S., Talbot C., Boutte Y., Catrice O., Read N.D., Satiat-Jeunemaitre B. (2004). FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J. Microsc. 214: 159–173 - PubMed
-
- Chen X., Irani N.G., Friml J. (2011). Clathrin-mediated endocytosis: The gateway into plant cells. Curr. Opin. Plant Biol. 14: 674–682 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

