What underlies the diversity of brain tumors?
- PMID: 23085857
- PMCID: PMC3586981
- DOI: 10.1007/s10555-012-9407-3
What underlies the diversity of brain tumors?
Abstract
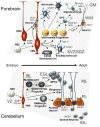
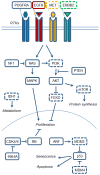
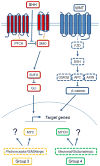
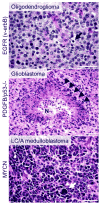
Glioma and medulloblastoma represent the most commonly occurring malignant brain tumors in adults and in children, respectively. Recent genomic and transcriptional approaches present a complex group of diseases and delineate a number of molecular subgroups within tumors that share a common histopathology. Differences in cells of origin, regional niches, developmental timing, and genetic events all contribute to this heterogeneity. In an attempt to recapitulate the diversity of brain tumors, an increasing array of genetically engineered mouse models (GEMMs) has been developed. These models often utilize promoters and genetic drivers from normal brain development and can provide insight into specific cells from which these tumors originate. GEMMs show promise in both developmental biology and developmental therapeutics. This review describes numerous murine brain tumor models in the context of normal brain development and the potential for these animals to impact brain tumor research.
Figures

References
-
- Lumsden A, Krumlauf R. Patterning the vertebrate neuraxis. Science. 1996;274(5290):1109–1115. - PubMed
-
- Liu A, Niswander LA. Bone morphogenetic protein signalling and vertebrate nervous system development. Nat Rev Neurosci. 2005;6(12):945–954. - PubMed
-
- Rowitch DH, Kriegstein AR. Developmental genetics of vertebrate glial-cell specification. Nature. 2010;468(7321):214–222. - PubMed
-
- Malatesta P, Hartfuss E, Gotz M. Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development. 2000;127(24):5253–5263. - PubMed
-
- Hansen DV, Lui JH, Parker PR, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464(7288):554–561. - PubMed
Publication types
MeSH terms
Grants and funding
- U54 CA163155/CA/NCI NIH HHS/United States
- R01 CA102321/CA/NCI NIH HHS/United States
- R01 CA133091/CA/NCI NIH HHS/United States
- CA128583/CA/NCI NIH HHS/United States
- P01 CA081403/CA/NCI NIH HHS/United States
- CA163155/CA/NCI NIH HHS/United States
- NS055750/NS/NINDS NIH HHS/United States
- CA148699/CA/NCI NIH HHS/United States
- CA081403/CA/NCI NIH HHS/United States
- R01 CA148699/CA/NCI NIH HHS/United States
- CA133091/CA/NCI NIH HHS/United States
- T32 CA128583/CA/NCI NIH HHS/United States
- R01 NS055750/NS/NINDS NIH HHS/United States
- CA102321/CA/NCI NIH HHS/United States

