Structure of the chemokine receptor CXCR1 in phospholipid bilayers
- PMID: 23086146
- PMCID: PMC3700570
- DOI: 10.1038/nature11580
Structure of the chemokine receptor CXCR1 in phospholipid bilayers
Abstract
CXCR1 is one of two high-affinity receptors for the CXC chemokine interleukin-8 (IL-8), a major mediator of immune and inflammatory responses implicated in many disorders, including tumour growth. IL-8, released in response to inflammatory stimuli, binds to the extracellular side of CXCR1. The ligand-activated intracellular signalling pathways result in neutrophil migration to the site of inflammation. CXCR1 is a class A, rhodopsin-like G-protein-coupled receptor (GPCR), the largest class of integral membrane proteins responsible for cellular signal transduction and targeted as drug receptors. Despite its importance, the molecular mechanism of CXCR1 signal transduction is poorly understood owing to the limited structural information available. Recent structural determination of GPCRs has advanced by modifying the receptors with stabilizing mutations, insertion of the protein T4 lysozyme and truncations of their amino acid sequences, as well as addition of stabilizing antibodies and small molecules that facilitate crystallization in cubic phase monoolein mixtures. The intracellular loops of GPCRs are crucial for G-protein interactions, and activation of CXCR1 involves both amino-terminal residues and extracellular loops. Our previous nuclear magnetic resonance studies indicate that IL-8 binding to the N-terminal residues is mediated by the membrane, underscoring the importance of the phospholipid bilayer for physiological activity. Here we report the three-dimensional structure of human CXCR1 determined by NMR spectroscopy. The receptor is in liquid crystalline phospholipid bilayers, without modification of its amino acid sequence and under physiological conditions. Features important for intracellular G-protein activation and signal transduction are revealed. The structure of human CXCR1 in a lipid bilayer should help to facilitate the discovery of new compounds that interact with GPCRs and combat diseases such as breast cancer.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

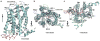
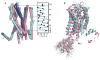
Comment in
-
G protein-coupled receptors: Spinning a native GPCR structure.Nat Rev Drug Discov. 2012 Dec;11(12):908. doi: 10.1038/nrd3900. Nat Rev Drug Discov. 2012. PMID: 23197037 No abstract available.
Similar articles
-
Interactions of interleukin-8 with the human chemokine receptor CXCR1 in phospholipid bilayers by NMR spectroscopy.J Mol Biol. 2011 Nov 25;414(2):194-203. doi: 10.1016/j.jmb.2011.08.025. Epub 2011 Oct 12. J Mol Biol. 2011. PMID: 22019593 Free PMC article.
-
Interaction of Monomeric Interleukin-8 with CXCR1 Mapped by Proton-Detected Fast MAS Solid-State NMR.Biophys J. 2017 Dec 19;113(12):2695-2705. doi: 10.1016/j.bpj.2017.09.041. Biophys J. 2017. PMID: 29262362 Free PMC article.
-
In silico analysis reveals sequential interactions and protein conformational changes during the binding of chemokine CXCL-8 to its receptor CXCR1.PLoS One. 2014 Apr 4;9(4):e94178. doi: 10.1371/journal.pone.0094178. eCollection 2014. PLoS One. 2014. PMID: 24705928 Free PMC article.
-
The CXCL8-CXCR1/2 pathways in cancer.Cytokine Growth Factor Rev. 2016 Oct;31:61-71. doi: 10.1016/j.cytogfr.2016.08.002. Epub 2016 Aug 25. Cytokine Growth Factor Rev. 2016. PMID: 27578214 Free PMC article. Review.
-
Structure determination of membrane proteins by nuclear magnetic resonance spectroscopy.Annu Rev Anal Chem (Palo Alto Calif). 2013;6:305-28. doi: 10.1146/annurev-anchem-062012-092631. Epub 2013 Apr 1. Annu Rev Anal Chem (Palo Alto Calif). 2013. PMID: 23577669 Free PMC article. Review.
Cited by
-
G Protein-Coupled Receptors (GPCRs) in Alzheimer's Disease: A Focus on BACE1 Related GPCRs.Front Aging Neurosci. 2016 Mar 24;8:58. doi: 10.3389/fnagi.2016.00058. eCollection 2016. Front Aging Neurosci. 2016. PMID: 27047374 Free PMC article. Review.
-
Structural Characterization of an LPA1 Second Extracellular Loop Mimetic with a Self-Assembling Coiled-Coil Folding Constraint.Int J Mol Sci. 2013 Jan 29;14(2):2788-807. doi: 10.3390/ijms14022788. Int J Mol Sci. 2013. PMID: 23434648 Free PMC article.
-
Boronic acid-containing aminopyridine- and aminopyrimidinecarboxamide CXCR1/2 antagonists: Optimization of aqueous solubility and oral bioavailability.Bioorg Med Chem Lett. 2015 Sep 15;25(18):3793-7. doi: 10.1016/j.bmcl.2015.07.090. Epub 2015 Jul 30. Bioorg Med Chem Lett. 2015. PMID: 26248802 Free PMC article.
-
Efficient and stable reconstitution of the ABC transporter BmrA for solid-state NMR studies.Front Mol Biosci. 2014 Jun 12;1:5. doi: 10.3389/fmolb.2014.00005. eCollection 2014. Front Mol Biosci. 2014. PMID: 25988146 Free PMC article.
-
Conserved Residues Control Activation of Mammalian G Protein-Coupled Odorant Receptors.J Am Chem Soc. 2015 Jul 8;137(26):8611-8616. doi: 10.1021/jacs.5b04659. Epub 2015 Jun 29. J Am Chem Soc. 2015. PMID: 26090619 Free PMC article.
References
-
- Holmes WE, Lee J, Kuang WJ, Rice GC, Wood WI. Structure and functional expression of a human interleukin-8 receptor. Science. 1991;253:1278–1280. - PubMed
-
- Sallusto F, Baggiolini M. Chemokines and leukocyte traffic. Nat Immunol. 2008;9:949–952. - PubMed
-
- Waugh DJ, Wilson C. The interleukin-8 pathway in cancer. Clin Cancer Res. 2008;14:6735–6741. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- P01 AI074805/AI/NIAID NIH HHS/United States
- R21 GM094727/GM/NIGMS NIH HHS/United States
- R01 EB005161/EB/NIBIB NIH HHS/United States
- R01GM075877/GM/NIGMS NIH HHS/United States
- R21 GM075917/GM/NIGMS NIH HHS/United States
- P41EB002031/EB/NIBIB NIH HHS/United States
- R21GM075917/GM/NIGMS NIH HHS/United States
- R21GM94727/GM/NIGMS NIH HHS/United States
- P41 EB002031/EB/NIBIB NIH HHS/United States
- R01 GM099986/GM/NIGMS NIH HHS/United States
- R01 GM075877/GM/NIGMS NIH HHS/United States
- P01AI074805/AI/NIAID NIH HHS/United States
- R01EB005161/EB/NIBIB NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

