Elf5 inhibits the epithelial-mesenchymal transition in mammary gland development and breast cancer metastasis by transcriptionally repressing Snail2
- PMID: 23086238
- PMCID: PMC3500637
- DOI: 10.1038/ncb2607
Elf5 inhibits the epithelial-mesenchymal transition in mammary gland development and breast cancer metastasis by transcriptionally repressing Snail2
Abstract
The epithelial-mesenchymal transition (EMT) is a complex process that occurs during organogenesis and in cancer metastasis. Despite recent progress, the molecular pathways connecting the physiological and pathological functions of EMT need to be better defined. Here we show that the transcription factor Elf5, a key regulator of mammary gland alveologenesis, controls EMT in both mammary gland development and metastasis. We uncovered this role for Elf5 through analyses of Elf5 conditional knockout animals, various in vitro and in vivo models of EMT and metastasis, an MMTV-neu transgenic model of mammary tumour progression and clinical breast cancer samples. Furthermore, we demonstrate that Elf5 suppresses EMT by directly repressing the transcription of Snail2, a master regulator of mammary stem cells and a known inducer of EMT. These findings establish Elf5 not only as a key cell lineage regulator during normal mammary gland development, but also as a suppressor of EMT and metastasis in breast cancer.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

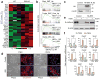



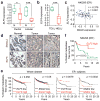


Comment in
-
Metastasis: Elf represses snail.Nat Rev Cancer. 2012 Dec;12(12):795. doi: 10.1038/nrc3406. Nat Rev Cancer. 2012. PMID: 23175114 No abstract available.
References
-
- Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. - PubMed
-
- Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. - PubMed
-
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

