Control of RelB during dendritic cell activation integrates canonical and noncanonical NF-κB pathways
- PMID: 23086447
- PMCID: PMC3634611
- DOI: 10.1038/ni.2446
Control of RelB during dendritic cell activation integrates canonical and noncanonical NF-κB pathways
Abstract
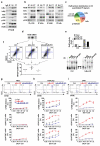
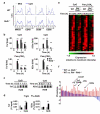
The NF-κB protein RelB controls dendritic cell (DC) maturation and may be targeted therapeutically to manipulate T cell responses in disease. Here we report that RelB promoted DC activation not as the expected RelB-p52 effector of the noncanonical NF-κB pathway, but as a RelB-p50 dimer regulated by canonical IκBs, IκBα and IκBɛ. IκB control of RelB minimized spontaneous maturation but enabled rapid pathogen-responsive maturation. Computational modeling of the NF-κB signaling module identified control points of this unexpected cell type-specific regulation. Fibroblasts that we engineered accordingly showed DC-like RelB control. Canonical pathway control of RelB regulated pathogen-responsive gene expression programs. This work illustrates the potential utility of systems analyses in guiding the development of combination therapeutics for modulating DC-dependent T cell responses.
Figures




Comment in
-
A less-canonical, canonical NF-κB pathway in DCs.Nat Immunol. 2012 Dec;13(12):1139-41. doi: 10.1038/ni.2476. Nat Immunol. 2012. PMID: 23160209 No abstract available.
References
-
- Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. - PubMed
-
- Liu YJ. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu Rev Immunol. 2005;23:275–306. - PubMed
-
- Serbina NV, Salazar-Mather TP, Biron CA, Kuziel WA, Pamer EG. TNF/iNOS-producing dendritic cells mediate innate immune defense against bacterial infection. Immunity. 2003;19:59–70. - PubMed
-
- Steinman RM, Banchereau J. Taking dendritic cells into medicine. Nature. 2007;449:419–426. - PubMed
-
- Carrasco D, Ryseck RP, Bravo R. Expression of relB transcripts during lymphoid organ development: specific expression in dendritic antigen-presenting cells. Development. 1993;118:1221–1231. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- GM071573/GM/NIGMS NIH HHS/United States
- GM085325/GM/NIGMS NIH HHS/United States
- P01 GM071862/GM/NIGMS NIH HHS/United States
- AI090935/AI/NIAID NIH HHS/United States
- AI081923/AI/NIAID NIH HHS/United States
- R01 AI083453/AI/NIAID NIH HHS/United States
- R01 AI081923/AI/NIAID NIH HHS/United States
- P50 GM085764/GM/NIGMS NIH HHS/United States
- R01 GM071573/GM/NIGMS NIH HHS/United States
- R01 GM085325/GM/NIGMS NIH HHS/United States
- GM085763/GM/NIGMS NIH HHS/United States
- P01 AI090935/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

