The CD46-Jagged1 interaction is critical for human TH1 immunity
- PMID: 23086448
- PMCID: PMC3505834
- DOI: 10.1038/ni.2454
The CD46-Jagged1 interaction is critical for human TH1 immunity
Abstract
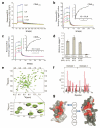
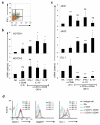
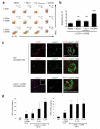
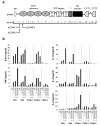
CD46 is a complement regulator with important roles related to the immune response. CD46 functions as a pathogen receptor and is a potent costimulator for the induction of interferon-γ (IFN-γ)-secreting effector T helper type 1 (T(H)1) cells and their subsequent switch into interleukin 10 (IL-10)-producing regulatory T cells. Here we identified the Notch family member Jagged1 as a physiological ligand for CD46. Furthermore, we found that CD46 regulated the expression of Notch receptors and ligands during T cell activation and that disturbance of the CD46-Notch crosstalk impeded induction of IFN-γ and switching to IL-10. Notably, CD4(+) T cells from CD46-deficient patients and patients with hypomorphic mutations in the gene encoding Jagged1 (Alagille syndrome) failed to mount appropriate T(H)1 responses in vitro and in vivo, which suggested that CD46-Jagged1 crosstalk is responsible for the recurrent infections in subpopulations of these patients.
Figures


Comment in
-
T cell responses: Jagged gives an edge to T(H)1 cells.Nat Rev Immunol. 2012 Dec;12(12):806-7. doi: 10.1038/nri3356. Nat Rev Immunol. 2012. PMID: 23175220 No abstract available.
References
-
- Liszewski MK, Post TW, Atkinson JP. Membrane cofactor protein (MCP or CD46): newest member of the regulators of complement activation gene cluster. Annu Rev Immunol. 1991;9:431–455. - PubMed
-
- Astier A, Trescol-Biemont MC, Azocar O, Lamouille B, Rabourdin-Combe C. Cutting edge: CD46, a new costimulatory molecule for T cells, that induces p120CBL and LAT phosphorylation. J Immunol. 2000;164:6091–6095. - PubMed
-
- Cope A, Le Friec G, Cardone J, Kemper C. The Th1 life cycle: molecular control of IFN-gamma to IL-10 switching. Trends Immunol. 2011;32:278–286. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

