Cell-free protein synthesis: the state of the art
- PMID: 23086573
- PMCID: PMC3553302
- DOI: 10.1007/s10529-012-1075-4
Cell-free protein synthesis: the state of the art
Abstract
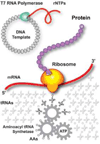
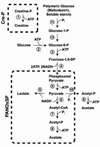
Cell-free protein synthesis harnesses the synthetic power of biology, programming the ribosomal translational machinery of the cell to create macromolecular products. Like PCR, which uses cellular replication machinery to create a DNA amplifier, cell-free protein synthesis is emerging as a transformative technology with broad applications in protein engineering, biopharmaceutical development, and post-genomic research. By breaking free from the constraints of cell-based systems, it takes the next step towards synthetic biology. Recent advances in reconstituted cell-free protein synthesis (Protein synthesis Using Recombinant Elements expression systems) are creating new opportunities to tailor the reactions for specialized applications including in vitro protein evolution, printing protein microarrays, isotopic labeling, and incorporating nonnatural amino acids.
Figures
Similar articles
-
Recent advances in development of cell-free protein synthesis systems for fast and efficient production of recombinant proteins.FEMS Microbiol Lett. 2018 Sep 1;365(17). doi: 10.1093/femsle/fny174. FEMS Microbiol Lett. 2018. PMID: 30084930 Review.
-
Improved cell-free RNA and protein synthesis system.PLoS One. 2014 Sep 2;9(9):e106232. doi: 10.1371/journal.pone.0106232. eCollection 2014. PLoS One. 2014. PMID: 25180701 Free PMC article.
-
Cell-free platforms for flexible expression and screening of enzymes.Biotechnol Adv. 2013 Nov;31(6):797-803. doi: 10.1016/j.biotechadv.2013.04.009. Epub 2013 May 4. Biotechnol Adv. 2013. PMID: 23648628 Review.
-
[Recent advances in cell-free unnatural protein synthesis].Sheng Wu Gong Cheng Xue Bao. 2018 Sep 25;34(9):1371-1385. doi: 10.13345/j.cjb.180203. Sheng Wu Gong Cheng Xue Bao. 2018. PMID: 30255672 Review. Chinese.
-
Applications of cell-free protein synthesis in synthetic biology: Interfacing bio-machinery with synthetic environments.Biotechnol J. 2013 Nov;8(11):1292-300. doi: 10.1002/biot.201200385. Epub 2013 Oct 4. Biotechnol J. 2013. PMID: 24123955 Review.
Cited by
-
Cell-free protein synthesis from a release factor 1 deficient Escherichia coli activates efficient and multiple site-specific nonstandard amino acid incorporation.ACS Synth Biol. 2014 Jun 20;3(6):398-409. doi: 10.1021/sb400140t. Epub 2014 Jan 2. ACS Synth Biol. 2014. PMID: 24328168 Free PMC article.
-
High-yield production of "difficult-to-express" proteins in a continuous exchange cell-free system based on CHO cell lysates.Sci Rep. 2017 Sep 15;7(1):11710. doi: 10.1038/s41598-017-12188-8. Sci Rep. 2017. PMID: 28916746 Free PMC article.
-
Unleashing the potential of mRNA therapeutics for inherited neurological diseases.Brain. 2024 Sep 3;147(9):2934-2945. doi: 10.1093/brain/awae135. Brain. 2024. PMID: 38662782 Free PMC article. Review.
-
Investigating and Optimizing the Lysate-Based Expression of Nonribosomal Peptide Synthetases Using a Reporter System.ACS Synth Biol. 2023 May 19;12(5):1447-1460. doi: 10.1021/acssynbio.2c00658. Epub 2023 Apr 11. ACS Synth Biol. 2023. PMID: 37039644 Free PMC article.
-
A Synopsis of Proteins and Their Purification.Methods Mol Biol. 2023;2699:1-14. doi: 10.1007/978-1-0716-3362-5_1. Methods Mol Biol. 2023. PMID: 37646990
References
-
- Beckert B, Masquida B. Synthesis of RNA by in vitro transcription. Methods Mol Biol. 2011;703:29–41. - PubMed
-
- Bremer H, Dennis PP. Chapter 5.2.3, Modulation of chemical composition and other parameters of the cell at different exponential growth rates. In: Böck A, Curtiss R III, Kaper JB, Karp PD, Neidhardt FC, Nyström T, Slauch JM, Squires CL, Ussery D, editors. EcoSal—Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, DC: ASM Press; 2008. http://www.ecosal.org. - PubMed
-
- Bundy BC, Swartz JR. Site-specific incorporation of p-propargyloxyphenylalanine in a cell-free environment for direct protein-protein click conjugation. Bioconjug Chem. 2010;21:255–263. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

