Neuron-restrictive silencer factor (NRSF) represses cocaine- and amphetamine-regulated transcript (CART) transcription and antagonizes cAMP-response element-binding protein signaling through a dual NRSE mechanism
- PMID: 23086924
- PMCID: PMC3522259
- DOI: 10.1074/jbc.M112.376590
Neuron-restrictive silencer factor (NRSF) represses cocaine- and amphetamine-regulated transcript (CART) transcription and antagonizes cAMP-response element-binding protein signaling through a dual NRSE mechanism
Abstract
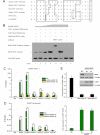
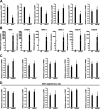
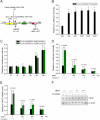
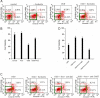
Cocaine- and amphetamine-regulated transcript (CART) peptide plays a pivotal role in neuroprotection against stroke-related brain injury. However, the regulatory mechanism on CART transcription, especially the repression mechanism, is not fully understood. Here, we show that the transcriptional repressor neuron-restrictive silencer elements (NRSF, also known as REST) represses CART expression through direct binding to two NRSF-binding elements (NRSEs) in the CART promoter and intron 1 (named pNRSE and iNRSE, respectively). EMSA show that NRSF binds to pNRSE and iNRSE directly in vitro. ChIP assays show that NRSF recruits differential co-repressor complexes including CoREST and HDAC1 to these NRSEs. The presence of both NRSEs is required for efficient repression of CART transcription as indicated by reporter gene assays. NRSF overexpression antagonizes forskolin-mediated up-regulation of CART mRNA and protein. Ischemia insult triggered by oxygen-glucose deprivation (OGD) enhances NRSF mRNA levels and then NRSF antagonizes the CREB signaling on CART activation, leading to augmented cell death. Depletion of NRSF in combination with forskolin treatment increases neuronal survival after ischemic insult. These findings reveal a novel dual NRSE mechanism by which NRSF represses CART expression and suggest that NRSF may serve as a therapeutic target for stroke treatment.
Figures




Similar articles
-
Regulatory role of neuron-restrictive silencing factor in the specific expression of cocaine- and amphetamine-regulated transcript gene.J Neurochem. 2008 Aug;106(3):1314-24. doi: 10.1111/j.1471-4159.2008.05487.x. Epub 2008 May 15. J Neurochem. 2008. PMID: 18485095
-
Transcriptional regulation of the neuropeptide VGF by the neuron-restrictive silencer factor/neuron-restrictive silencer element.Neuroreport. 2015 Feb 11;26(3):144-51. doi: 10.1097/WNR.0000000000000316. Neuroreport. 2015. PMID: 25569790
-
Transcriptional repression by neuron-restrictive silencer factor is mediated via the Sin3-histone deacetylase complex.Mol Cell Biol. 2000 Mar;20(6):2147-57. doi: 10.1128/MCB.20.6.2147-2157.2000. Mol Cell Biol. 2000. PMID: 10688661 Free PMC article.
-
Brain REST/NRSF Is Not Only a Silent Repressor but Also an Active Protector.Mol Neurobiol. 2017 Jan;54(1):541-550. doi: 10.1007/s12035-015-9658-4. Epub 2016 Jan 7. Mol Neurobiol. 2017. PMID: 26742529 Review.
-
Regulation of the cholinergic gene locus by the repressor element-1 silencing transcription factor/neuron restrictive silencer factor (REST/NRSF).Life Sci. 2004 Mar 19;74(18):2213-25. doi: 10.1016/j.lfs.2003.08.045. Life Sci. 2004. PMID: 15017977 Review.
Cited by
-
ChIP-Seq Data Mining: Remarkable Differences in NRSF/REST Target Genes between Human ESC and ESC-Derived Neurons.Bioinform Biol Insights. 2013 Dec 1;7:357-68. doi: 10.4137/BBI.S13279. eCollection 2013. Bioinform Biol Insights. 2013. PMID: 24324330 Free PMC article.
-
Haploinsufficiency of X-linked intellectual disability gene CASK induces post-transcriptional changes in synaptic and cellular metabolic pathways.Exp Neurol. 2020 Jul;329:113319. doi: 10.1016/j.expneurol.2020.113319. Epub 2020 Apr 17. Exp Neurol. 2020. PMID: 32305418 Free PMC article.
-
CART treatment improves memory and synaptic structure in APP/PS1 mice.Sci Rep. 2015 May 11;5:10224. doi: 10.1038/srep10224. Sci Rep. 2015. PMID: 25959573 Free PMC article.
-
REST Is Not Resting: REST/NRSF in Health and Disease.Biomolecules. 2023 Oct 2;13(10):1477. doi: 10.3390/biom13101477. Biomolecules. 2023. PMID: 37892159 Free PMC article. Review.
-
Targeting histone deacetylases: perspectives for epigenetic-based therapy in cardio-cerebrovascular disease.J Geriatr Cardiol. 2015 Mar;12(2):153-64. doi: 10.11909/j.issn.1671-5411.2015.02.010. J Geriatr Cardiol. 2015. PMID: 25870619 Free PMC article. Review.
References
-
- Kuhar M. J., Dall Vechia S. E. (1999) CART peptides. Novel addiction- and feeding-related neuropeptides. Trends Neurosci. 22, 316–320 - PubMed
-
- Kuriyama G., Takekoshi S., Tojo K., Nakai Y., Kuhar M. J., Osamura R. Y. (2004) Cocaine- and amphetamine-regulated transcript peptide in the rat anterior pituitary gland is localized in gonadotrophs and suppresses prolactin secretion. Endocrinology 145, 2542–2550 - PubMed
-
- Vicentic A., Dominguez G., Hunter R. G., Philpot K., Wilson M., Kuhar M. J. (2004) Cocaine- and amphetamine-regulated transcript peptide levels in blood exhibit a diurnal rhythm. Regulation by glucocorticoids. Endocrinology 145, 4119–4124 - PubMed
-
- Upadhya M. A., Nakhate K. T., Kokare D. M., Singh U., Singru P. S., Subhedar N. K. (2012) CART peptide in the nucleus accumbens shell acts downstream to dopamine and mediates the reward and reinforcement actions of morphine. Neuropharmacology 62, 1823–1833 - PubMed
-
- Balkan B., Keser A., Gozen O., Koylu E. O., Dagci T., Kuhar M. J., Pogun S. (2012) Forced swim stress elicits region-specific changes in CART expression in the stress axis and stress regulatory brain areas. Brain Res. 1432, 56–65 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

