Structural basis for sirtuin activity and inhibition
- PMID: 23086949
- PMCID: PMC3522243
- DOI: 10.1074/jbc.R112.372300
Structural basis for sirtuin activity and inhibition
Abstract
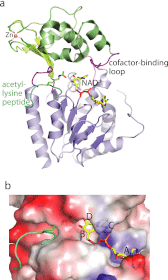
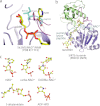
Sir2 proteins, or sirtuins, are a family of enzymes that catalyze NAD(+)-dependent deacetylation reactions and can also process ribosyltransferase, demalonylase, and desuccinylase activities. More than 40 crystal structures of sirtuins have been determined, alone or in various liganded forms. These high-resolution architectural details lay the foundation for understanding the molecular mechanisms of catalysis, regulation, substrate specificity, and inhibition of sirtuins. In this minireview, we summarize these structural features and discuss their implications for understanding sirtuin function.
Figures

References
-
- Bell S. D., Botting C. H., Wardleworth B. N., Jackson S. P., White M. F. (2002) The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science 296, 148–151 - PubMed
-
- Starai V. J., Celic I., Cole R. N., Boeke J. D., Escalante-Semerena J. C. (2002) Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science 298, 2390–2392 - PubMed
-
- Kim E. J., Um S. J. (2008) SIRT1: roles in aging and cancer. BMB Rep. 41, 751–756 - PubMed
-
- Guarente L. (2007) Sirtuins in aging and disease. Cold Spring Harb. Symp. Quant. Biol. 72, 483–488 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

