Human Rap1 interacts directly with telomeric DNA and regulates TRF2 localization at the telomere
- PMID: 23086976
- PMCID: PMC3516710
- DOI: 10.1074/jbc.M112.415984
Human Rap1 interacts directly with telomeric DNA and regulates TRF2 localization at the telomere
Abstract
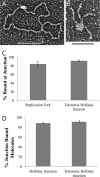
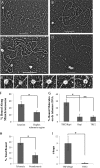
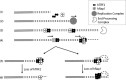
The TRF2-Rap1 complex suppresses non-homologous end joining and interacts with DNAPK-C to prevent end joining. We previously demonstrated that hTRF2 is a double strand telomere binding protein that forms t-loops in vitro and recognizes three- and four-way junctions independent of DNA sequence. How the DNA binding characteristics of hTRF2 to DNA is altered in the presence of hRap1 however is not known. Here we utilized EM and quantitative gel retardation to characterize the DNA binding properties of hRap1 and the TRF2-Rap1 complex. Both gel filtration chromatography and mass analysis from two-dimensional projections showed that the TRF2-Rap1 complex exists in solution and binds to DNA as a complex consisting of four monomers each of hRap1 and hTRF2. EM revealed for the first time that hRap1 binds to DNA templates in the absence of hTRF2 with a preference for double strand-single strand junctions in a sequence independent manner. When hTRF2 and hRap1 are in a complex, its affinity for ds telomeric sequences is 2-fold higher than TRF2 alone and more than 10-fold higher for telomeric 3' ends. This suggests that as hTRF2 recruits hRap1 to telomeric sequences, hRap1 alters the affinity of hTRF2 and its binding preference on telomeric DNA. Moreover, the TRF2-Rap1 complex has higher ability to re-model telomeric DNA than either component alone. This finding underlies the importance of complex formation between hRap1 and hTRF2 for telomere function and end protection.
Figures



References
-
- Griffith J. D., Comeau L., Rosenfield S., Stansel R. M., Bianchi A., Moss H., de Lange T. (1999) Mammalian telomeres end in a large duplex loop. Cell 97, 503–514 - PubMed
-
- Liu D., O'Connor M. S., Qin J., Songyang Z. (2004) Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J. Biol. Chem. 279, 51338–51342 - PubMed
-
- Palm W., de Lange T. (2008) How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42, 301–334 - PubMed
-
- Zhu X. D., Niedernhofer L., Kuster B., Mann M., Hoeijmakers J. H., de Lange T. (2003) ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol. Cell 12, 1489–1498 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

