Amyloid β-sheet mimics that antagonize protein aggregation and reduce amyloid toxicity
- PMID: 23089868
- PMCID: PMC3481199
- DOI: 10.1038/nchem.1433
Amyloid β-sheet mimics that antagonize protein aggregation and reduce amyloid toxicity
Abstract
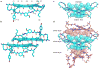
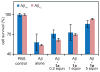
The amyloid protein aggregation associated with diseases such as Alzheimer's, Parkinson's and type II diabetes (among many others) features a bewildering variety of β-sheet-rich structures in transition from native proteins to ordered oligomers and fibres. The variation in the amino-acid sequences of the β-structures presents a challenge to developing a model system of β-sheets for the study of various amyloid aggregates. Here, we introduce a family of robust β-sheet macrocycles that can serve as a platform to display a variety of heptapeptide sequences from different amyloid proteins. We have tailored these amyloid β-sheet mimics (ABSMs) to antagonize the aggregation of various amyloid proteins, thereby reducing the toxicity of amyloid aggregates. We describe the structures and inhibitory properties of ABSMs containing amyloidogenic peptides from the amyloid-β peptide associated with Alzheimer's disease, β(2)-microglobulin associated with dialysis-related amyloidosis, α-synuclein associated with Parkinson's disease, islet amyloid polypeptide associated with type II diabetes, human and yeast prion proteins, and Tau, which forms neurofibrillary tangles.
Conflict of interest statement
The authors declare no conflict.
Figures

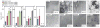
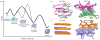
References
-
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. - PubMed
-
- Aguzzi A, O’Connor T. Protein aggregation diseases: pathogenicity and therapeutic perspectives. Nat Rev Drug Discov. 2010;9:237–248. - PubMed
-
- Bartolini M, Andrisano V. Strategies for the inhibition of protein aggregation in human diseases. ChemBioChem. 2010;11:1018–1035. - PubMed
-
- Greenwald J, Riek R. Biology of amyloid: structure, function, and regulation. Structure. 2010;18:1244–1260. - PubMed
Publication types
MeSH terms
Substances
Associated data
- PubChem-Substance/137292014
- PubChem-Substance/137292015
- PubChem-Substance/137292016
- PubChem-Substance/137292017
- PubChem-Substance/137292018
- PubChem-Substance/137292019
- PubChem-Substance/137292020
- PubChem-Substance/137292021
- PubChem-Substance/137292022
- PubChem-Substance/137292023
- PubChem-Substance/137292024
- PubChem-Substance/137292025
- PubChem-Substance/137292026
- PubChem-Substance/137292027
- PubChem-Substance/137292028
- PubChem-Substance/137292029
- PubChem-Substance/137292030
- PubChem-Substance/137292031
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

