Molecular classification of gliomas based on whole genome gene expression: a systematic report of 225 samples from the Chinese Glioma Cooperative Group
- PMID: 23090983
- PMCID: PMC3499016
- DOI: 10.1093/neuonc/nos263
Molecular classification of gliomas based on whole genome gene expression: a systematic report of 225 samples from the Chinese Glioma Cooperative Group
Abstract
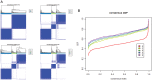
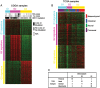
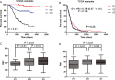
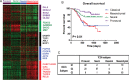
Defining glioma subtypes based on objective genetic and molecular signatures may allow for a more rational, patient-specific approach to molecularly targeted therapy. However, prior studies attempting to classify glioma subtypes have given conflicting results. We aim to complement and validate the existing molecular classification system on a large number of samples from an East Asian population. A total of 225 samples from Chinese patients was selected for whole genome gene expression profiling. Consensus clustering was applied. Three major groups of gliomas were identified (referred to as G1, G2, and G3). The G1 subgroup correlates with a good clinical outcome, young age, and extremely high frequency of IDH1 mutations. Relative to the G1 subgroup, the G3 subgroup is correlated with a poorer clinical outcome, older age, and a very low rate of mutations in the IDH1 gene. Correlations of the G2 subgroup with respect to clinical outcome, age, and IDH1 mutation fall between the G1 and G3 subgroups. In addition, the G2 subtype was associated with a higher percentage of loss of 1p/19q when compared with G1 and G3 subtypes. Furthermore, our classification scheme was validated on 2 independent datasets derived from the cancer genome atlas (TCGA) and Rembrandt. With use of the TCGA classification system, proneural, neural, and mesenchymal, but not classical subtype, associated gene signatures were clearly defined. In summary, our results reveal that 3 main subtypes stably exist in Chinese patients with glioma. Our classification scheme may reflect the clinical and genetic alterations more clearly. Classical subtype-associated gene signature was not found in our dataset.
Figures
References
-
- Yan W, Zhang W, Jiang T. Oncogene addiction in gliomas: implications for molecular targeted therapy. J Exp Clin Cancer Res. 2011;30:58. doi:10.1186/1756-9966-30-58. - DOI - PMC - PubMed
-
- Drappatz J, Norden AD, Wen PY. Therapeutic strategies for inhibiting invasion in glioblastoma. Expert Rev Neurother. 2009;9(4):519–534. doi:10.1586/ern.09.10. - DOI - PubMed
-
- Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi:10.1016/j.ccr.2009.12.020. - DOI - PMC - PubMed
-
- Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi:10.1016/j.ccr.2006.02.019. - DOI - PubMed
-
- Li A, Walling J, Ahn S, et al. Unsupervised analysis of transcriptomic profiles reveals six glioma subtypes. Cancer Res. 2009;69(5):2091–2099. doi:10.1158/0008-5472.CAN-08-2100. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

