MDA5 assembles into a polar helical filament on dsRNA
- PMID: 23090998
- PMCID: PMC3494895
- DOI: 10.1073/pnas.1212186109
MDA5 assembles into a polar helical filament on dsRNA
Abstract
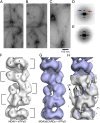
Melanoma differentiation-associated protein 5 (MDA5) detects viral dsRNA in the cytoplasm. On binding of RNA, MDA5 forms polymers, which trigger assembly of the signaling adaptor mitochondrial antiviral-signaling protein (MAVS) into its active fibril form. The molecular mechanism of MDA5 signaling is not well understood, however. Here we show that MDA5 forms helical filaments on dsRNA and report the 3D structure of the filaments using electron microscopy (EM) and image reconstruction. MDA5 assembles into a polar, single-start helix around the RNA. Fitting of an MDA5 homology model into the structure suggests a key role for the MDA5 C-terminal domain in cooperative filament assembly. Our study supports a signal transduction mechanism in which the helical array of MDA5 within filaments nucleates the assembly of MAVS fibrils. We conclude that MDA5 is a polymerization-dependent signaling platform that uses the amyloid-like self-propagating properties of MAVS to amplify signaling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441(7089):101–105. - PubMed
-
- Hornung V, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314(5801):994–997. - PubMed
-
- Pichlmair A, et al. RIG-I–mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314(5801):997–1001. - PubMed
-
- Kowalinski E, et al. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147(2):423–435. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

