Histone H3K4 demethylation is negatively regulated by histone H3 acetylation in Saccharomyces cerevisiae
- PMID: 23091032
- PMCID: PMC3494965
- DOI: 10.1073/pnas.1202070109
Histone H3K4 demethylation is negatively regulated by histone H3 acetylation in Saccharomyces cerevisiae
Abstract
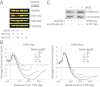
Histone H3 lysine 4 trimethylation (H3K4me3) is a hallmark of transcription initiation, but how H3K4me3 is demethylated during gene repression is poorly understood. Jhd2, a JmjC domain protein, was recently identified as the major H3K4me3 histone demethylase (HDM) in Saccharomyces cerevisiae. Although JHD2 is required for removal of methylation upon gene repression, deletion of JHD2 does not result in increased levels of H3K4me3 in bulk histones, indicating that this HDM is unable to demethylate histones during steady-state conditions. In this study, we showed that this was due to the negative regulation of Jhd2 activity by histone H3 lysine 14 acetylation (H3K14ac), which colocalizes with H3K4me3 across the yeast genome. We demonstrated that loss of the histone H3-specific acetyltransferases (HATs) resulted in genome-wide depletion of H3K4me3, and this was not due to a transcription defect. Moreover, H3K4me3 levels were reestablished in HAT mutants following loss of JHD2, which suggested that H3-specific HATs and Jhd2 serve opposing functions in regulating H3K4me3 levels. We revealed the molecular basis for this suppression by demonstrating that H3K14ac negatively regulated Jhd2 demethylase activity on an acetylated peptide in vitro. These results revealed the existence of a general mechanism for removal of H3K4me3 following gene repression.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


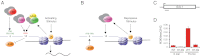
References
-
- Van Holde KE. Chromatin. New York: Springer; 1989.
-
- Morillon A, Karabetsou N, Nair A, Mellor J. Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol Cell. 2005;18(6):723–734. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

