Cellular origin and pathophysiology of chronic lymphocytic leukemia
- PMID: 23091163
- PMCID: PMC3501361
- DOI: 10.1084/jem.20120833
Cellular origin and pathophysiology of chronic lymphocytic leukemia
Abstract
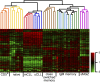
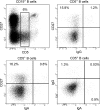
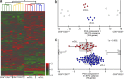
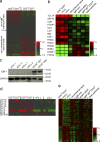
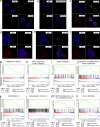
The cellular origin of chronic lymphocytic leukemia (CLL) is still debated, although this information is critical to understanding its pathogenesis. Transcriptome analyses of CLL and the main normal B cell subsets from human blood and spleen revealed that immunoglobulin variable region (IgV) gene unmutated CLL derives from unmutated mature CD5(+) B cells and mutated CLL derives from a distinct, previously unrecognized CD5(+)CD27(+) post-germinal center B cell subset. Stereotyped V gene rearrangements are enriched among CD5(+) B cells, providing independent evidence for a CD5(+) B cell derivation of CLL. Notably, these CD5(+) B cell populations include oligoclonal expansions already found in young healthy adults, putatively representing an early phase in CLL development before the CLL precursor lesion monoclonal B cell lymphocytosis. Finally, we identified deregulated proteins, including EBF1 and KLF transcription factors, that were not detected in previous comparisons of CLL and conventional B cells.
Figures

References
-
- Allman D., Jain A., Dent A., Maile R.R., Selvaggi T., Kehry M.R., Staudt L.M. 1996. BCL-6 expression during B-cell activation. Blood. 87:5257–5268 - PubMed
-
- Andréasson U., Edén P., Peterson C., Högerkorp C.M., Jerkeman M., Andersen N., Berglund M., Sundström C., Rosenquist R., Borrebaeck C.A., Ek S. 2010. Identification of uniquely expressed transcription factors in highly purified B-cell lymphoma samples. Am. J. Hematol. 85:418–425 - PubMed
-
- Bachl J., Carlson C., Gray-Schopfer V., Dessing M., Olsson C. 2001. Increased transcription levels induce higher mutation rates in a hypermutating cell line. J. Immunol. 166:5051–5057 - PubMed
-
- Barrena S., Almeida J., Yunta M., López A., Fernández-Mosteirín N., Giralt M., Romero M., Perdiguer L., Delgado M., Orfao A., Lazo P.A. 2005. Aberrant expression of tetraspanin molecules in B-cell chronic lymphoproliferative disorders and its correlation with normal B-cell maturation. Leukemia. 19:1376–1383 10.1038/sj.leu.2403822 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

