Small sizes and indolent evolutionary dynamics challenge the potential role of P2RY8-CRLF2-harboring clones as main relapse-driving force in childhood ALL
- PMID: 23091296
- PMCID: PMC4194314
- DOI: 10.1182/blood-2012-07-443218
Small sizes and indolent evolutionary dynamics challenge the potential role of P2RY8-CRLF2-harboring clones as main relapse-driving force in childhood ALL
Erratum in
- Blood. 2013 Aug 15;122(7):1328
Abstract
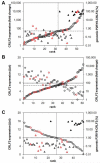
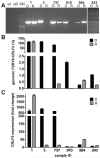
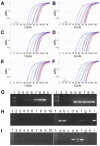
The P2RY8-CRLF2 fusion defines a particular relapse-prone subset of childhood acute lymphoblastic leukemia (ALL) in Italian Association of Pediatric Hematology and Oncology Berlin-Frankfurt-Münster (AIEOP-BFM) 2000 protocols. To investigate whether and to what extent different clone sizes influence disease and relapse development, we quantified the genomic P2RY8-CRLF2 fusion product and correlated it with the corresponding CRLF2 expression levels in patients enrolled in the BFM-ALL 2000 protocol in Austria. Of 268 cases without recurrent chromosomal translocations and high hyperdiploidy, representing approximately 50% of all cases, 67 (25%) were P2RY8-CRLF2 positive. The respective clone sizes were ≥ 20% in 27% and < 20% in 73% of them. The cumulative incidence of relapse of the entire fusion-positive group was clone size independent and significantly higher than that of the fusion-negative group (35% ± 8% vs 13% ± 3%, P = .008) and primarily confined to the non-high-risk group. Of 22 P2RY8-CRLF2-positive diagnosis/relapse pairs, only 4/8 had the fusion-positive dominant clone conserved at relapse, whereas none of the original 14 fusion-positive small clones reappeared as the dominant relapse clone. We conclude that the majority of P2RY8-CRLF2-positive clones are small at diagnosis and virtually never generate a dominant relapse clone. Our findings therefore suggest that P2RY8-CRLF2-positive clones do not have the necessary proliferative or selective advantage to evolve into a disease-relevant relapse clone.
Figures


References
-
- Cario G, Zimmermann M, Romey R, et al. Presence of the P2RY8-CRLF2 rearrangement is associated with a poor prognosis in non-high-risk precursor B-cell acute lymphoblastic leukemia in children treated according to the ALL-BFM 2000 protocol. Blood. 2010;115(26):5393–5397. - PubMed
-
- Russell LJ, Capasso M, Vater I, et al. Deregulated expression of cytokine receptor gene, CRLF2, is involved in lymphoid transformation in B-cell precursor acute lymphoblastic leukemia. Blood. 2009;114(13):2688–2698. - PubMed
-
- Ensor HM, Schwab C, Russell LJ, et al. Demographic, clinical, and outcome features of children with acute lymphoblastic leukemia and CRLF2 deregulation: results from the MRC ALL97 clinical trial. Blood. 2011;117(7):2129–2136. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

