Mycoplasma pneumoniae large DNA repetitive elements RepMP1 show type specific organization among strains
- PMID: 23091634
- PMCID: PMC3472980
- DOI: 10.1371/journal.pone.0047625
Mycoplasma pneumoniae large DNA repetitive elements RepMP1 show type specific organization among strains
Abstract
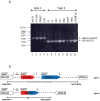
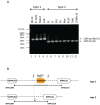
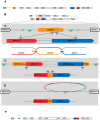
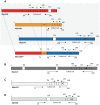
Mycoplasma pneumoniae is the smallest self-replicating bacterium with a streamlined genome of 0.81 Mb. Complete genome analysis revealed the presence of multiple copies of four large repetitive elements (designated RepMP1, RepMP2/3, RepMP4 and RepMP5) that are implicated in creating sequence variations among individual strains. Recently, we described RepMP1-associated sequence variations between reference strain M129 and clinical isolate S1 that involved three RepMP1-genes (i.e. mpn130, mpn137 and mpn138). Using PCR and sequencing we analyze 28 additional M. pneumoniae strains and demonstrate the existence of S1-like sequence variants in nine strains and M129-like variants in the remaining nineteen strains. We propose a series of recombination steps that facilitates transition from M129- to S1-like sequence variants. Next we examined the remaining RepMP1-genes and observed no other rearrangements related to the repeat element. The only other detected difference was varying numbers of the 21-nucleotide tandem repeats within mpn127, mpn137, mpn501 and mpn524. Furthermore, typing of strains through analysis of large RepMPs localized within the adhesin P1 operon revealed that sequence divergence involving RepMP1-genes mpn130, mpn137 and mpn138 is strictly type-specific. Once more our analysis confirmed existence of two highly conserved groups of M. pneumoniae strains.
Conflict of interest statement
Figures
References
-
- Baseman JB (1993) The cytadhesins of Mycoplasma pneumoniae and M. genitalium . Subcell Biochem 20: 243–259. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

