Akt phosphorylates the transcriptional repressor bmi1 to block its effects on the tumor-suppressing ink4a-arf locus
- PMID: 23092893
- PMCID: PMC3784651
- DOI: 10.1126/scisignal.2003199
Akt phosphorylates the transcriptional repressor bmi1 to block its effects on the tumor-suppressing ink4a-arf locus
Abstract
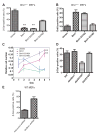
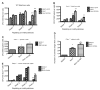
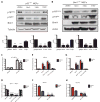
The Polycomb group protein Bmi1 is a transcriptional silencer of the Ink4a-Arf locus, which encodes the cell cycle regulator p16(Ink4a) and the tumor suppressor p19(Arf). Bmi1 plays a key role in oncogenesis and stem cell self-renewal. We report that phosphorylation of human Bmi1 at Ser³¹⁶ by Akt impaired its function by triggering its dissociation from the Ink4a-Arf locus, which resulted in decreased ubiquitylation of histone H2A and the inability of Bmi1 to promote cellular proliferation and tumor growth. Moreover, Akt-mediated phosphorylation of Bmi1 also inhibited its ability to promote self-renewal of hematopoietic stem and progenitor cells. Our study provides a mechanism for the increased abundance of p16(Ink4a) and p19(Arf) seen in cancer cells with an activated phosphoinositide 3-kinase to Akt signaling pathway and identifies crosstalk between phosphorylation events and chromatin structure.
Conflict of interest statement
Figures

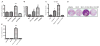


Similar articles
-
A novel zinc finger protein Zfp277 mediates transcriptional repression of the Ink4a/arf locus through polycomb repressive complex 1.PLoS One. 2010 Aug 24;5(8):e12373. doi: 10.1371/journal.pone.0012373. PLoS One. 2010. PMID: 20808772 Free PMC article.
-
Bmi1 promotes hepatic stem cell expansion and tumorigenicity in both Ink4a/Arf-dependent and -independent manners in mice.Hepatology. 2010 Sep;52(3):1111-23. doi: 10.1002/hep.23793. Hepatology. 2010. PMID: 20648475
-
Polycomb mediated epigenetic silencing and replication timing at the INK4a/ARF locus during senescence.PLoS One. 2009 May 20;4(5):e5622. doi: 10.1371/journal.pone.0005622. PLoS One. 2009. PMID: 19462008 Free PMC article.
-
Epigenetic regulation of the INK4b-ARF-INK4a locus: in sickness and in health.Epigenetics. 2010 Nov-Dec;5(8):685-90. doi: 10.4161/epi.5.8.12996. Epub 2010 Nov 1. Epigenetics. 2010. PMID: 20716961 Free PMC article. Review.
-
A protein synthesis brake for hematopoietic stem cell maintenance.Genes Dev. 2022 Aug 1;36(15-16):871-873. doi: 10.1101/gad.350107.122. Genes Dev. 2022. PMID: 36207141 Free PMC article. Review.
Cited by
-
Radiosensitization of Primary Human Glioblastoma Stem-like Cells with Low-Dose AKT Inhibition.Mol Cancer Ther. 2015 May;14(5):1171-80. doi: 10.1158/1535-7163.MCT-14-0708. Epub 2015 Feb 18. Mol Cancer Ther. 2015. PMID: 25695954 Free PMC article.
-
The prognostic significance of BMI1 expression in invasive breast cancer is dependent on its molecular subtypes.Breast Cancer Res Treat. 2020 Aug;182(3):581-589. doi: 10.1007/s10549-020-05719-x. Epub 2020 Jun 10. Breast Cancer Res Treat. 2020. PMID: 32524353 Free PMC article.
-
Mitogen-activated protein kinase signaling mediates phosphorylation of polycomb ortholog Cbx7.J Biol Chem. 2013 Dec 20;288(51):36398-408. doi: 10.1074/jbc.M113.486266. Epub 2013 Nov 5. J Biol Chem. 2013. PMID: 24194518 Free PMC article.
-
Akt-mediated phosphorylation controls the activity of the Y-box protein MSY3 in skeletal muscle.Skelet Muscle. 2015 May 29;5:18. doi: 10.1186/s13395-015-0043-9. eCollection 2015. Skelet Muscle. 2015. PMID: 26146542 Free PMC article.
-
The Crucial Roles of Bmi-1 in Cancer: Implications in Pathogenesis, Metastasis, Drug Resistance, and Targeted Therapies.Int J Mol Sci. 2022 Jul 26;23(15):8231. doi: 10.3390/ijms23158231. Int J Mol Sci. 2022. PMID: 35897796 Free PMC article. Review.
References
-
- Valk-Lingbeek ME, Bruggeman SW, van Lohuizen M. Stem cells and cancer: The Polycomb connection. Cell. 2004;118:409–418. - PubMed
-
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. - PubMed
-
- Pietersen AM, van Lohuizen M. Stem cell regulation by polycomb repressors: Postponing commitment. Curr Opin Cell Biol. 2008;20:201–207. - PubMed
-
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. - PubMed
-
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

