X-inactivation: quantitative predictions of protein interactions in the Xist network
- PMID: 23093590
- PMCID: PMC3592426
- DOI: 10.1093/nar/gks968
X-inactivation: quantitative predictions of protein interactions in the Xist network
Abstract
The transcriptional silencing of one of the female X-chromosomes is a finely regulated process that requires accumulation in cis of the long non-coding RNA X-inactive-specific transcript (Xist) followed by a series of epigenetic modifications. Little is known about the molecular machinery regulating initiation and maintenance of chromosomal silencing. Here, we introduce a new version of our algorithm catRAPID to investigate Xist associations with a number of proteins involved in epigenetic regulation, nuclear scaffolding, transcription and splicing processes. Our method correctly identifies binding regions and affinities of protein interactions, providing a powerful theoretical framework for the study of X-chromosome inactivation and other events mediated by ribonucleoprotein associations.
Figures
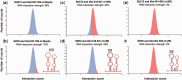

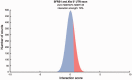



References
-
- Navarro P, Avner P. An embryonic story: analysis of the gene regulative network controlling Xist expression in mouse embryonic stem cells. Bioessays. 2010;32:581–588. - PubMed
-
- Tattermusch A, Brockdorff N. A scaffold for X chromosome inactivation. Hum. Genet. 2011;130:247–253. - PubMed
-
- Wutz A. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat. Rev. Genet. 2011;12:542–553. - PubMed
-
- Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat. Genet. 1999;21:400–404. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

