Exploring Ty1 retrotransposon RNA structure within virus-like particles
- PMID: 23093595
- PMCID: PMC3592414
- DOI: 10.1093/nar/gks983
Exploring Ty1 retrotransposon RNA structure within virus-like particles
Abstract
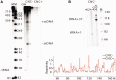
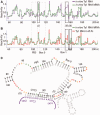
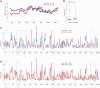
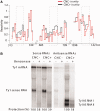
Ty1, a long terminal repeat retrotransposon of Saccharomyces, is structurally and functionally related to retroviruses. However, a differentiating aspect between these retroelements is the diversity of the replication strategies used by long terminal repeat retrotransposons. To understand the structural organization of cis-acting elements present on Ty1 genomic RNA from the GAG region that control reverse transcription, we applied chemoenzymatic probing to RNA/tRNA complexes assembled in vitro and to the RNA in virus-like particles. By comparing different RNA states, our analyses provide a comprehensive structure of the primer-binding site, a novel pseudoknot adjacent to the primer-binding sites, three regions containing palindromic sequences that may be involved in RNA dimerization or packaging and candidate protein interaction sites. In addition, we determined the impact of a novel form of transposon control based on Ty1 antisense transcripts that associate with virus-like particles. Our results support the idea that antisense RNAs inhibit retrotransposition by targeting Ty1 protein function rather than annealing with the RNA genome.
Figures

Similar articles
-
Retroviral-like determinants and functions required for dimerization of Ty1 retrotransposon RNA.RNA Biol. 2019 Dec;16(12):1749-1763. doi: 10.1080/15476286.2019.1657370. Epub 2019 Aug 30. RNA Biol. 2019. PMID: 31469343 Free PMC article.
-
Structure-Function Model for Kissing Loop Interactions That Initiate Dimerization of Ty1 RNA.Viruses. 2017 Apr 26;9(5):93. doi: 10.3390/v9050093. Viruses. 2017. PMID: 28445416 Free PMC article.
-
Retrotransposon Ty1 RNA contains a 5'-terminal long-range pseudoknot required for efficient reverse transcription.RNA. 2013 Mar;19(3):320-32. doi: 10.1261/rna.035535.112. Epub 2013 Jan 17. RNA. 2013. PMID: 23329695 Free PMC article.
-
Determinants of Genomic RNA Encapsidation in the Saccharomyces cerevisiae Long Terminal Repeat Retrotransposons Ty1 and Ty3.Viruses. 2016 Jul 14;8(7):193. doi: 10.3390/v8070193. Viruses. 2016. PMID: 27428991 Free PMC article. Review.
-
tRNAs as primer of reverse transcriptases.Biochimie. 1995;77(1-2):113-24. doi: 10.1016/0300-9084(96)88114-4. Biochimie. 1995. PMID: 7541250 Review.
Cited by
-
Mapping the structural landscape of the yeast Ty3 retrotransposon RNA genome.Nucleic Acids Res. 2024 Sep 9;52(16):9821-9837. doi: 10.1093/nar/gkae494. Nucleic Acids Res. 2024. PMID: 38864374 Free PMC article.
-
Ty1 escapes restriction by the self-encoded factor p22 through mutations in capsid.Mob Genet Elements. 2016 Mar 7;6(2):e1154639. doi: 10.1080/2159256X.2016.1154639. eCollection 2016 Mar-Apr. Mob Genet Elements. 2016. PMID: 27141327 Free PMC article.
-
RNAthor - fast, accurate normalization, visualization and statistical analysis of RNA probing data resolved by capillary electrophoresis.PLoS One. 2020 Oct 1;15(10):e0239287. doi: 10.1371/journal.pone.0239287. eCollection 2020. PLoS One. 2020. PMID: 33002005 Free PMC article.
-
The Ty1 LTR-retrotransposon of budding yeast, Saccharomyces cerevisiae.Microbiol Spectr. 2015 Apr 1;3(2):1-35. doi: 10.1128/microbiolspec.MDNA3-0053-2014. Microbiol Spectr. 2015. PMID: 25893143 Free PMC article.
-
Co-translational localization of an LTR-retrotransposon RNA to the endoplasmic reticulum nucleates virus-like particle assembly sites.PLoS Genet. 2014 Mar 6;10(3):e1004219. doi: 10.1371/journal.pgen.1004219. eCollection 2014 Mar. PLoS Genet. 2014. PMID: 24603646 Free PMC article.
References
-
- Voytas DF, Boeke JD. Ty1 and Ty5 of Saccharomyces cerevisiae. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington, DC: ASM Press; 2002. pp. 614–630.
-
- Lesage P, Todeschini A. Happy together: the life and times of Ty retrotransposons and their hosts. Cytogenet. Genome Res. 2005;110:70–90. - PubMed

