The Comparative Toxicogenomics Database: update 2013
- PMID: 23093600
- PMCID: PMC3531134
- DOI: 10.1093/nar/gks994
The Comparative Toxicogenomics Database: update 2013
Abstract
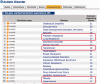
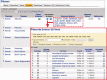
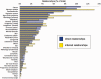
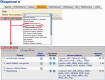
The Comparative Toxicogenomics Database (CTD; http://ctdbase.org/) provides information about interactions between environmental chemicals and gene products and their relationships to diseases. Chemical-gene, chemical-disease and gene-disease interactions manually curated from the literature are integrated to generate expanded networks and predict many novel associations between different data types. CTD now contains over 15 million toxicogenomic relationships. To navigate this sea of data, we added several new features, including DiseaseComps (which finds comparable diseases that share toxicogenomic profiles), statistical scoring for inferred gene-disease and pathway-chemical relationships, filtering options for several tools to refine user analysis and our new Gene Set Enricher (which provides biological annotations that are enriched for gene sets). To improve data visualization, we added a Cytoscape Web view to our ChemComps feature, included color-coded interactions and created a 'slim list' for our MEDIC disease vocabulary (allowing diseases to be grouped for meta-analysis, visualization and better data management). CTD continues to promote interoperability with external databases by providing content and cross-links to their sites. Together, this wealth of expanded chemical-gene-disease data, combined with novel ways to analyze and view content, continues to help users generate testable hypotheses about the molecular mechanisms of environmental diseases.
Figures


Similar articles
-
The Comparative Toxicogenomics Database: update 2011.Nucleic Acids Res. 2011 Jan;39(Database issue):D1067-72. doi: 10.1093/nar/gkq813. Epub 2010 Sep 22. Nucleic Acids Res. 2011. PMID: 20864448 Free PMC article.
-
The Comparative Toxicogenomics Database: update 2017.Nucleic Acids Res. 2017 Jan 4;45(D1):D972-D978. doi: 10.1093/nar/gkw838. Epub 2016 Sep 19. Nucleic Acids Res. 2017. PMID: 27651457 Free PMC article.
-
The Comparative Toxicogenomics Database: update 2019.Nucleic Acids Res. 2019 Jan 8;47(D1):D948-D954. doi: 10.1093/nar/gky868. Nucleic Acids Res. 2019. PMID: 30247620 Free PMC article.
-
The Comparative Toxicogenomics Database (CTD): a resource for comparative toxicological studies.J Exp Zool A Comp Exp Biol. 2006 Sep 1;305(9):689-92. doi: 10.1002/jez.a.307. J Exp Zool A Comp Exp Biol. 2006. PMID: 16902965 Free PMC article. Review.
-
Chemical databases for environmental health and clinical research.Toxicol Lett. 2009 Apr 10;186(1):62-5. doi: 10.1016/j.toxlet.2008.10.003. Epub 2008 Oct 18. Toxicol Lett. 2009. PMID: 18996453 Free PMC article. Review.
Cited by
-
A modularity-based method reveals mixed modules from chemical-gene heterogeneous network.PLoS One. 2015 Apr 30;10(4):e0125585. doi: 10.1371/journal.pone.0125585. eCollection 2015. PLoS One. 2015. PMID: 25927435 Free PMC article.
-
PsyGeNET: a knowledge platform on psychiatric disorders and their genes.Bioinformatics. 2015 Sep 15;31(18):3075-7. doi: 10.1093/bioinformatics/btv301. Epub 2015 May 11. Bioinformatics. 2015. PMID: 25964630 Free PMC article.
-
Enabling Web-scale data integration in biomedicine through Linked Open Data.NPJ Digit Med. 2019 Sep 10;2:90. doi: 10.1038/s41746-019-0162-5. eCollection 2019. NPJ Digit Med. 2019. PMID: 31531395 Free PMC article. Review.
-
The Rat Genome Database 2015: genomic, phenotypic and environmental variations and disease.Nucleic Acids Res. 2015 Jan;43(Database issue):D743-50. doi: 10.1093/nar/gku1026. Epub 2014 Oct 29. Nucleic Acids Res. 2015. PMID: 25355511 Free PMC article.
-
How consistent are we? Interlaboratory comparison study in fathead minnows using the model estrogen 17α-ethinylestradiol to develop recommendations for environmental transcriptomics.Environ Toxicol Chem. 2017 Oct;36(10):2614-2623. doi: 10.1002/etc.3799. Epub 2017 Apr 19. Environ Toxicol Chem. 2017. PMID: 28316117 Free PMC article.
References
-
- Mortensen HM, Euling SY. Integrating mechanistic and polymorphism data to characterize human genetic susceptiblity for environmental chemical risk assessment in the 21st centruy. Toxicol. Appl. Pharmacol. 2011 February 1. (doi:10.1016/j.taap.2011.01.015; epub ahead of print) - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

