The Online Protein Processing Resource (TOPPR): a database and analysis platform for protein processing events
- PMID: 23093603
- PMCID: PMC3531153
- DOI: 10.1093/nar/gks998
The Online Protein Processing Resource (TOPPR): a database and analysis platform for protein processing events
Abstract
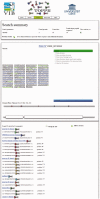
We here present The Online Protein Processing Resource (TOPPR; http://iomics.ugent.be/toppr/), an online database that contains thousands of published proteolytically processed sites in human and mouse proteins. These cleavage events were identified with COmbinded FRActional DIagonal Chromatography proteomics technologies, and the resulting database is provided with full data provenance. Indeed, TOPPR provides an interactive visual display of the actual fragmentation mass spectrum that led to each identification of a reported processed site, complete with fragment ion annotations and search engine scores. Apart from warehousing and disseminating these data in an intuitive manner, TOPPR also provides an online analysis platform, including methods to analyze protease specificity and substrate-centric analyses. Concretely, TOPPR supports three ways to retrieve data: (i) the retrieval of all substrates for one or more cellular stimuli or assays; (ii) a substrate search by UniProtKB/Swiss-Prot accession number, entry name or description; and (iii) a motif search that retrieves substrates matching a user-defined protease specificity profile. The analysis of the substrates is supported through the presence of a variety of annotations, including predicted secondary structure, known domains and experimentally obtained 3D structure where available. Across substrates, substrate orthologs and conserved sequence stretches can also be shown, with iceLogo visualization provided for the latter.
Figures
References
-
- Puente XS, Sánchez LM, Overall CM, López-Otín C. Human and mouse proteases: a comparative genomic approach. Nat. Rev. Genet. 2003;4:544–548. - PubMed
-
- Gevaert K, Goethals M, Martens L, Van Damme J, Staes A, Thomas G, Vandekerckhove J. Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat. Biotechnol. 2003;21:566–569. - PubMed
-
- Van Damme P, Martens L, Van Damme J, Hugelier K, Staes A, Vandekerckhove J, Gevaert K. Caspase-specific and nonspecific in vivo protein processing during Fas-induced apoptosis. Nat. Methods. 2005;2:771–777. - PubMed
-
- Van Damme P, Staes A, Bronsoms S, Helsens K, Colaert N, Timmerman E, Aviles FX, Vandekerckhove J, Gevaert K. Complementary positional proteomics for screening substrates of endo- and exoproteases. Nat. Methods. 2010;7:512–515. - PubMed
-
- Vande Walle L, Van Damme P, Lamkanfi M, Saelens X, Vandekerckhove J, Gevaert K, Vandenabeele P. Proteome-wide identification of HtrA2/Omi substrates. J. Proteome Res. 2007;6:1006–1015. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

