Comparison of mRNA features affecting translation initiation and reinitiation
- PMID: 23093605
- PMCID: PMC3592434
- DOI: 10.1093/nar/gks989
Comparison of mRNA features affecting translation initiation and reinitiation
Abstract
Regulation of gene expression at the level of translation accounts for up to three orders of magnitude in its efficiency. We systematically compared the impact of several mRNA features on translation initiation at the first gene in an operon with those for the second gene. Experiments were done in a system with internal control based on dual cerulean and red (CER/RFP) fluorescent proteins. We demonstrated significant differences in the efficiency of Shine Dalgarno sequences acting at the leading gene and at the following genes in an operon. The majority of frequent intercistronic arrangements possess medium SD dependence, medium dependence on the preceding cistron translation and efficient stimulation by A/U-rich sequences. The second cistron starting immediately after preceding cistron stop codon displays unusually high dependence on the SD sequence.
Figures

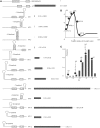




References
-
- Dreyfus M. What constitutes the signal for the initiation of protein synthesis on Escherichia coli mRNAs? J. Mol. Biol. 1988;204:79–94. - PubMed
-
- Butler JS, Springer M, Dondon J, Graffe M, Grunberg-Manago M. Escherichia coli protein synthesis initiation factor IF3 controls its own gene expression at the translational level in vivo. J. Mol. Biol. 1986;192:767–780. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

