Bacterial and fungal chitinase chiJ orthologs evolve under different selective constraints following horizontal gene transfer
- PMID: 23095575
- PMCID: PMC3506478
- DOI: 10.1186/1756-0500-5-581
Bacterial and fungal chitinase chiJ orthologs evolve under different selective constraints following horizontal gene transfer
Abstract
Background: Certain bacteria from the genus Streptomyces are currently used as biological control agents against plant pathogenic fungi. Hydrolytic enzymes that degrade fungal cell wall components, such as chitinases, are suggested as one possible mechanism in biocontrol interactions. Adaptive evolution of chitinases are previously reported for plant chitinases involved in defence against fungal pathogens, and in fungal chitinases involved in fungal-fungal interactions. In this study we investigated the molecular evolution of chitinase chiJ in the bacterial genus Streptomyces. In addition, as chiJ orthologs are previously reported in certain fungal species as a result from horizontal gene transfer, we conducted a comparative study of differences in evolutionary patterns between bacterial and fungal taxa.
Findings: ChiJ contained three sites evolving under strong positive selection and four groups of co-evolving sites. Regions of high amino acid diversity were predicted to be surface-exposed and associated with coil regions that connect certain α-helices and β-strands in the family 18 chitinase TIM barrel structure, but not associated with the catalytic cleft. The comparative study with fungal ChiJ orthologs identified three regions that display signs of type 1 functional divergence, where unique adaptations in the bacterial and fungal taxa are driven by positive selection.
Conclusions: The identified surface-exposed regions of chitinase ChiJ where sequence diversification is driven by positive selection may putatively be related to functional divergence between bacterial and fungal orthologs. These results show that ChiJ orthologs have evolved under different selective constraints following the horizontal gene transfer event.
Figures
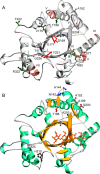

References
-
- Tews I, van Scheltinga ACT, Perrakis A, Wilson KS, Dijkstra BW. Substrate-assisted catalysis unifies two families of chitinolytic enzymes. J Am Chem Soc. 1997;119:7954–7959. doi: 10.1021/ja970674i. - DOI
-
- Hodgson DA. Primary metabolism and its control in Streptomycetes: A most unusual group of bacteria. Adv Microb Physiol. 2000;42:47–238. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical

