3D-FISH analysis of embryonic nuclei in mouse highlights several abrupt changes of nuclear organization during preimplantation development
- PMID: 23095683
- PMCID: PMC3517311
- DOI: 10.1186/1471-213X-12-30
3D-FISH analysis of embryonic nuclei in mouse highlights several abrupt changes of nuclear organization during preimplantation development
Abstract
Background: Embryonic development proceeds through finely tuned reprogramming of the parental genomes to form a totipotent embryo. Cells within this embryo will then differentiate and give rise to all the tissues of a new individual. Early embryonic development thus offers a particularly interesting system in which to analyze functional nuclear organization. When the organization of higher-order chromatin structures, such as pericentromeric heterochromatin, was first analyzed in mouse embryos, specific nuclear rearrangements were observed that correlated with embryonic genome activation at the 2-cell stage. However, most existing analyses have been conducted by visual observation of fluorescent images, in two dimensions or on z-stack sections/projections, but only rarely in three dimensions (3D).
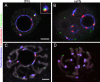
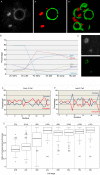
Results: In the present study, we used DNA fluorescent in situ hybridization (FISH) to localize centromeric (minor satellites), pericentromeric (major satellites), and telomeric genomic sequences throughout the preimplantation period in naturally fertilized mouse embryos (from the 1-cell to blastocyst stage). Their distribution was then analyzed in 3D on confocal image stacks, focusing on the nucleolar precursor bodies and nucleoli known to evolve rapidly throughout the first developmental stages. We used computational imaging to quantify various nuclear parameters in the 3D-FISH images, to analyze the organization of compartments of interest, and to measure physical distances between these compartments.
Conclusions: The results highlight differences in nuclear organization between the two parental inherited genomes at the 1-cell stage, i.e. just after fertilization. We also found that the reprogramming of the embryonic genome, which starts at the 2-cell stage, undergoes other remarkable changes during preimplantation development, particularly at the 4-cell stage.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

