Analysis of composition-based metagenomic classification
- PMID: 23095761
- PMCID: PMC3477002
- DOI: 10.1186/1471-2164-13-S5-S1
Analysis of composition-based metagenomic classification
Abstract
Background: An essential step of a metagenomic study is the taxonomic classification, that is, the identification of the taxonomic lineage of the organisms in a given sample. The taxonomic classification process involves a series of decisions. Currently, in the context of metagenomics, such decisions are usually based on empirical studies that consider one specific type of classifier. In this study we propose a general framework for analyzing the impact that several decisions can have on the classification problem. Instead of focusing on any specific classifier, we define a generic score function that provides a measure of the difficulty of the classification task. Using this framework, we analyze the impact of the following parameters on the taxonomic classification problem: (i) the length of n-mers used to encode the metagenomic sequences, (ii) the similarity measure used to compare sequences, and (iii) the type of taxonomic classification, which can be conventional or hierarchical, depending on whether the classification process occurs in a single shot or in several steps according to the taxonomic tree.
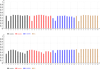
Results: We defined a score function that measures the degree of separability of the taxonomic classes under a given configuration induced by the parameters above. We conducted an extensive computational experiment and found out that reasonable values for the parameters of interest could be (i) intermediate values of n, the length of the n-mers; (ii) any similarity measure, because all of them resulted in similar scores; and (iii) the hierarchical strategy, which performed better in all of the cases.
Conclusions: As expected, short n-mers generate lower configuration scores because they give rise to frequency vectors that represent distinct sequences in a similar way. On the other hand, large values for n result in sparse frequency vectors that represent differently metagenomic fragments that are in fact similar, also leading to low configuration scores. Regarding the similarity measure, in contrast to our expectations, the variation of the measures did not change the configuration scores significantly. Finally, the hierarchical strategy was more effective than the conventional strategy, which suggests that, instead of using a single classifier, one should adopt multiple classifiers organized as a hierarchy.
Figures

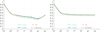
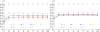

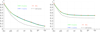

Similar articles
-
Deep learning models for bacteria taxonomic classification of metagenomic data.BMC Bioinformatics. 2018 Jul 9;19(Suppl 7):198. doi: 10.1186/s12859-018-2182-6. BMC Bioinformatics. 2018. PMID: 30066629 Free PMC article.
-
MyTaxa: an advanced taxonomic classifier for genomic and metagenomic sequences.Nucleic Acids Res. 2014 Apr;42(8):e73. doi: 10.1093/nar/gku169. Epub 2014 Mar 3. Nucleic Acids Res. 2014. PMID: 24589583 Free PMC article.
-
INDUS - a composition-based approach for rapid and accurate taxonomic classification of metagenomic sequences.BMC Genomics. 2011 Nov 30;12 Suppl 3(Suppl 3):S4. doi: 10.1186/1471-2164-12-S3-S4. Epub 2011 Nov 30. BMC Genomics. 2011. PMID: 22369237 Free PMC article.
-
Benchmarking Metagenomics Tools for Taxonomic Classification.Cell. 2019 Aug 8;178(4):779-794. doi: 10.1016/j.cell.2019.07.010. Cell. 2019. PMID: 31398336 Free PMC article. Review.
-
Classification of metagenomic sequences: methods and challenges.Brief Bioinform. 2012 Nov;13(6):669-81. doi: 10.1093/bib/bbs054. Epub 2012 Sep 8. Brief Bioinform. 2012. PMID: 22962338 Review.
Cited by
-
Evaluation of shotgun metagenomics sequence classification methods using in silico and in vitro simulated communities.BMC Bioinformatics. 2015 Nov 4;16:363. doi: 10.1186/s12859-015-0788-5. BMC Bioinformatics. 2015. PMID: 26537885 Free PMC article.
-
A Robust Framework for Microbial Archaeology.Annu Rev Genomics Hum Genet. 2017 Aug 31;18:321-356. doi: 10.1146/annurev-genom-091416-035526. Epub 2017 Apr 26. Annu Rev Genomics Hum Genet. 2017. PMID: 28460196 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

