Sex-biased transcriptome evolution in Drosophila
- PMID: 23097318
- PMCID: PMC3514954
- DOI: 10.1093/gbe/evs093
Sex-biased transcriptome evolution in Drosophila
Abstract
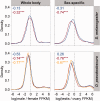
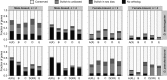
Sex-biased genes are thought to drive phenotypic differences between males and females. The recent availability of high-throughput gene expression data for many related species has led to a burst of investigations into the genomic and evolutionary properties of sex-biased genes. In Drosophila, a number of studies have found that X chromosomes are deficient in male-biased genes (demasculinized) and enriched for female-biased genes (feminized) and that male-biased genes evolve faster than female-biased genes. However, studies have yielded vastly different conclusions regarding the numbers of sex-biased genes and forces shaping their evolution. Here, we use RNA-seq data from multiple tissues of Drosophila melanogaster and D. pseudoobscura, a species with a recently evolved X chromosome, to explore the evolution of sex-biased genes in Drosophila. First, we compare several independent metrics for classifying sex-biased genes and find that the overlap of genes identified by different metrics is small, particularly for female-biased genes. Second, we investigate genome-wide expression patterns and uncover evidence of demasculinization and feminization of both ancestral and new X chromosomes, demonstrating that gene content on sex chromosomes evolves rapidly. Third, we examine the evolutionary rates of sex-biased genes and show that male-biased genes evolve much faster than female-biased genes, which evolve at similar rates to unbiased genes. Analysis of gene expression among tissues reveals that this trend may be partially due to pleiotropic effects of female-biased genes, which limits their evolutionary potential. Thus, our findings illustrate the importance of accurately identifying sex-biased genes and provide insight into their evolutionary dynamics in Drosophila.
Figures







References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;5:403–410. - PubMed
-
- Anders S. Analyzing RNA-Seq data with the “DESeq” package. Mol Biol. 2010;43:1–17.
-
- Beckenbach AT, Wei YW, Liu H. Relationships in the Drosophila obscura species group, inferred from mitochondrial cytochrome oxidase II sequences. Mol Biol Evol. 1993;10:619–634. - PubMed
-
- Connallon T, Knowles L. Intergenomic conflict revealed by patterns of sex-biased gene expression. Trends Genet. 2005;21:495–499. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

