Alternative polyadenylation: new insights from global analyses
- PMID: 23097429
- PMCID: PMC3504663
- DOI: 10.1261/rna.035899.112
Alternative polyadenylation: new insights from global analyses
Abstract
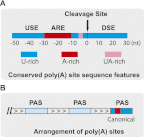
Recent studies have revealed widespread mRNA alternative polyadenylation (APA) in eukaryotes and its dynamic spatial and temporal regulation. APA not only generates proteomic and functional diversity, but also plays important roles in regulating gene expression. Global deregulation of APA has been demonstrated in a variety of human diseases. Recent exciting advances in the field have been made possible in a large part by high throughput analyses using newly developed experimental tools. Here I review the recent progress in global studies of APA and the insights that have emerged from these and other studies that use more conventional methods.
Figures

References
-
- Alt FW, Bothwell AL, Knapp M, Siden E, Mather E, Koshland M, Baltimore D 1980. Synthesis of secreted and membrane-bound immunoglobulin μ heavy chains is directed by mRNAs that differ at their 3′ ends. Cell 20: 293–301 - PubMed
-
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A 2004. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432: 112–118 - PubMed
-
- Bennett CL, Brunkow ME, Ramsdell F, O'Briant KC, Zhu Q, Fuleihan RL, Shigeoka AO, Ochs HD, Chance PF 2001. A rare polyadenylation signal mutation of the FOXP3 gene (AAUAAA→AAUGAA) leads to the IPEX syndrome. Immunogenetics 53: 435–439 - PubMed
-
- Bentley DL 2005. Rules of engagement: Co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol 17: 251–256 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
