Transcriptomic profiling of the oyster pathogen Vibrio splendidus opens a window on the evolutionary dynamics of the small RNA repertoire in the Vibrio genus
- PMID: 23097430
- PMCID: PMC3504672
- DOI: 10.1261/rna.033324.112
Transcriptomic profiling of the oyster pathogen Vibrio splendidus opens a window on the evolutionary dynamics of the small RNA repertoire in the Vibrio genus
Abstract
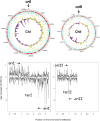
Work in recent years has led to the recognition of the importance of small regulatory RNAs (sRNAs) in bacterial regulation networks. New high-throughput sequencing technologies are paving the way to the exploration of an expanding sRNA world in nonmodel bacteria. In the Vibrio genus, compared to the enterobacteriaceae, still a limited number of sRNAs have been characterized, mostly in Vibrio cholerae, where they have been shown to be important for virulence, as well as in Vibrio harveyi. In addition, genome-wide approaches in V. cholerae have led to the discovery of hundreds of potential new sRNAs. Vibrio splendidus is an oyster pathogen that has been recently associated with massive mortality episodes in the French oyster growing industry. Here, we report the first RNA-seq study in a Vibrio outside of the V. cholerae species. We have uncovered hundreds of candidate regulatory RNAs, be it cis-regulatory elements, antisense RNAs, and trans-encoded sRNAs. Conservation studies showed the majority of them to be specific to V. splendidus. However, several novel sRNAs, previously unidentified, are also present in V. cholerae. Finally, we identified 28 trans sRNAs that are conserved in all the Vibrio genus species for which a complete genome sequence is available, possibly forming a Vibrio "sRNA core."
Figures




Similar articles
-
csrB Gene Duplication Drives the Evolution of Redundant Regulatory Pathways Controlling Expression of the Major Toxic Secreted Metalloproteases in Vibrio tasmaniensis LGP32.mSphere. 2018 Nov 28;3(6):e00582-18. doi: 10.1128/mSphere.00582-18. mSphere. 2018. PMID: 30487156 Free PMC article.
-
Transcriptomic Approaches for Studying Quorum Sensing in Vibrio cholerae.Methods Enzymol. 2018;612:303-342. doi: 10.1016/bs.mie.2018.09.008. Epub 2018 Oct 24. Methods Enzymol. 2018. PMID: 30502947
-
Modulation of Vibrio cholerae gene expression through conjugative delivery of engineered regulatory small RNAs.J Bacteriol. 2024 Oct 24;206(10):e0014224. doi: 10.1128/jb.00142-24. Epub 2024 Sep 18. J Bacteriol. 2024. PMID: 39292012 Free PMC article.
-
Non-coding sRNAs regulate virulence in the bacterial pathogen Vibrio cholerae.RNA Biol. 2012 Apr;9(4):392-401. doi: 10.4161/rna.19975. Epub 2012 Apr 1. RNA Biol. 2012. PMID: 22546941 Free PMC article. Review.
-
Small RNAs in streptococci.RNA Biol. 2012 Apr;9(4):414-26. doi: 10.4161/rna.20104. Epub 2012 Apr 1. RNA Biol. 2012. PMID: 22546939 Review.
Cited by
-
The ilvGMEDA Operon Is Regulated by Transcription Attenuation in Vibrio alginolyticus ZJ-T.Appl Environ Microbiol. 2019 Sep 17;85(19):e00880-19. doi: 10.1128/AEM.00880-19. Print 2019 Oct 1. Appl Environ Microbiol. 2019. PMID: 31324637 Free PMC article.
-
Meta-omics approaches reveal unique small RNAs exhibited by the uncultured microorganisms dwelling deep-sea hydrothermal sediment in Guaymas Basin.Arch Microbiol. 2022 Jul 6;204(8):461. doi: 10.1007/s00203-022-03085-4. Arch Microbiol. 2022. PMID: 35792953
-
[Regulatory role of small RNA srn821978 in mutacin IV expression in Streptococcus mutans].Nan Fang Yi Ke Da Xue Xue Bao. 2021 Nov 20;41(11):1725-1732. doi: 10.12122/j.issn.1673-4254.2021.11.19. Nan Fang Yi Ke Da Xue Xue Bao. 2021. PMID: 34916201 Free PMC article. Chinese.
-
Impact of bacterial sRNAs in stress responses.Biochem Soc Trans. 2017 Dec 15;45(6):1203-1212. doi: 10.1042/BST20160363. Epub 2017 Nov 3. Biochem Soc Trans. 2017. PMID: 29101308 Free PMC article. Review.
-
Carrageenan catabolism is encoded by a complex regulon in marine heterotrophic bacteria.Nat Commun. 2017 Nov 22;8(1):1685. doi: 10.1038/s41467-017-01832-6. Nat Commun. 2017. PMID: 29162826 Free PMC article.
References
-
- Alix E, Blanc-Potard AB 2009. Hydrophobic peptides: Novel regulators within bacterial membrane. Mol Microbiol 72: 5–11 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
