APOBEC3G restricts HIV-1 to a greater extent than APOBEC3F and APOBEC3DE in human primary CD4+ T cells and macrophages
- PMID: 23097438
- PMCID: PMC3536366
- DOI: 10.1128/JVI.00676-12
APOBEC3G restricts HIV-1 to a greater extent than APOBEC3F and APOBEC3DE in human primary CD4+ T cells and macrophages
Abstract
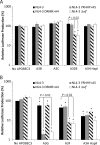
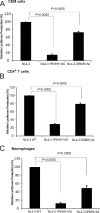
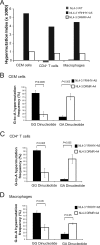
APOBEC3 proteins inhibit HIV-1 replication in experimental systems and induce hypermutation in infected patients; however, the relative contributions of several APOBEC3 proteins to restriction of HIV-1 replication in the absence of the viral Vif protein in human primary CD4(+) T cells and macrophages are unknown. We observed significant inhibition of HIV-1Δvif produced in 293T cells in the presence of APOBEC3DE (A3DE), APOBEC3F (A3F), APOBEC3G (A3G), and APOBEC3H haplotype II (A3H HapII) but not APOBEC3B (A3B), APOBEC3C (A3C), or APOBEC3H haplotype I (A3H HapI). Our previous studies showed that Vif amino acids Y(40)RHHY(44) are important for inducing proteasomal degradation of A3G, whereas amino acids (14)DRMR(17) are important for degradation of A3F and A3DE. Here, we introduced substitution mutations of (40)YRHHY(44) and (14)DRMR(17) in replication-competent HIV-1 to generate vif mutants NL4-3 YRHHY>A5 and NL4-3 DRMR>A4 to compare the antiviral activity of A3G to the combined antiviral activity of A3F and A3DE in activated CD4(+) T cells and macrophages. During the first 15 days (round 1), in which multiple cycles of viral replication occurred, both the NL4-3 YRHHY>A5 and NL4-3 DRMR>A4 mutants replicated in activated CD4(+) T cells and macrophages, and only the NL4-3 YRHHY>A5 mutant showed a 2- to 4-day delay in replication compared to the wild type. During the subsequent 27 days (round 2) of cultures initiated with peak virus obtained from round 1, the NL4-3 YRHHY>A5 mutant exhibited a longer, 8- to 10-day delay and the NL4-3 DRMR>A4 mutant exhibited a 2- to 6-day delay in replication compared to the wild type. The NL4-3 YRHHY>A5 and NL4-3 DRMR>A4 mutant proviruses displayed G-to-A hypermutations primarily in GG and GA dinucleotides as expected of A3G- and A3F- or A3DE-mediated deamination, respectively. We conclude that A3G exerts a greater restriction effect on HIV-1 than A3F and A3DE.
Figures

References
-
- Liddament MT, Brown WL, Schumacher AJ, Harris RS. 2004. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 14:1385–1391 - PubMed
-
- Rose KM, Marin M, Kozak SL, Kabat D. 2005. Regulated production and anti-HIV type 1 activities of cytidine deaminases APOBEC3B, 3F, and 3G. AIDS Res. Hum. Retroviruses 21:611–619 - PubMed
-
- Sheehy AM, Gaddis NC, Choi JD, Malim MH. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646–650 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

