Mutations in multiple domains of Gag drive the emergence of in vitro resistance to the phosphonate-containing HIV-1 protease inhibitor GS-8374
- PMID: 23097440
- PMCID: PMC3536386
- DOI: 10.1128/JVI.01211-12
Mutations in multiple domains of Gag drive the emergence of in vitro resistance to the phosphonate-containing HIV-1 protease inhibitor GS-8374
Abstract
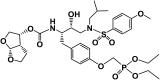
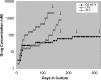
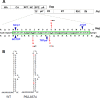
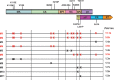
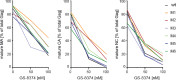
GS-8374 is a potent HIV protease inhibitor (PI) with a unique diethyl-phosphonate moiety. Due to a balanced contribution of enthalpic and entropic components to its interaction with the protease (PR) active site, the compound retains activity against HIV mutants with high-level multi-PI resistance. We report here the in vitro selection and characterization of HIV variants resistant to GS-8374. While highly resistant viruses with multiple mutations in PR were isolated in the presence of control PIs, an HIV variant displaying moderate (14-fold) resistance to GS-8374 was generated only after prolonged passaging for >300 days. The isolate showed low-level cross-resistance to darunavir, atazanavir, lopinavir, and saquinavir, but not other PIs, and contained a single R41K mutation in PR combined with multiple genotypic changes in the Gag matrix, capsid, nucleocapsid, and SP2 domains. Mutations also occurred in the transframe peptide and p6* domain of the Gag-Pol polyprotein. Analysis of recombinant HIV variants indicated that mutations in Gag, but not the R41K in PR, conferred reduced susceptibility to GS-8374. The Gag mutations acted in concert, since they did not affect susceptibility when introduced individually. Analysis of viral particles revealed that the mutations rendered Gag more susceptible to PR-mediated cleavage in the presence of GS-8374. In summary, the emergence of resistance to GS-8374 involved a combination of substrate mutations without typical resistance mutations in PR. These substrate changes were distributed throughout Gag and acted in an additive manner. Thus, they are classified as primary resistance mutations indicating a unique mechanism and pathway of resistance development for GS-8374.
Figures
Similar articles
-
In Vivo Emergence of a Novel Protease Inhibitor Resistance Signature in HIV-1 Matrix.mBio. 2020 Nov 3;11(6):e02036-20. doi: 10.1128/mBio.02036-20. mBio. 2020. PMID: 33144375 Free PMC article.
-
Non-cleavage site gag mutations in amprenavir-resistant human immunodeficiency virus type 1 (HIV-1) predispose HIV-1 to rapid acquisition of amprenavir resistance but delay development of resistance to other protease inhibitors.J Virol. 2009 Apr;83(7):3059-68. doi: 10.1128/JVI.02539-08. Epub 2009 Jan 28. J Virol. 2009. PMID: 19176623 Free PMC article.
-
Positive selection pressure introduces secondary mutations at Gag cleavage sites in human immunodeficiency virus type 1 harboring major protease resistance mutations.J Virol. 2009 Sep;83(17):8916-24. doi: 10.1128/JVI.00003-09. Epub 2009 Jun 10. J Virol. 2009. PMID: 19515784 Free PMC article.
-
Reviewing HIV-1 Gag Mutations in Protease Inhibitors Resistance: Insights for Possible Novel Gag Inhibitor Designs.Molecules. 2019 Sep 6;24(18):3243. doi: 10.3390/molecules24183243. Molecules. 2019. PMID: 31489889 Free PMC article. Review.
-
Darunavir: a review of its use in the management of HIV infection in adults.Drugs. 2009;69(4):477-503. doi: 10.2165/00003495-200969040-00007. Drugs. 2009. PMID: 19323590 Review.
Cited by
-
In Vivo Emergence of a Novel Protease Inhibitor Resistance Signature in HIV-1 Matrix.mBio. 2020 Nov 3;11(6):e02036-20. doi: 10.1128/mBio.02036-20. mBio. 2020. PMID: 33144375 Free PMC article.
-
A sensitive assay using a native protein substrate for screening HIV-1 maturation inhibitors targeting the protease cleavage site between the matrix and capsid.Biochemistry. 2013 Jul 23;52(29):4929-40. doi: 10.1021/bi4005232. Epub 2013 Jul 11. Biochemistry. 2013. PMID: 23763575 Free PMC article.
-
HIV Genome-Wide Protein Associations: a Review of 30 Years of Research.Microbiol Mol Biol Rev. 2016 Jun 29;80(3):679-731. doi: 10.1128/MMBR.00065-15. Print 2016 Sep. Microbiol Mol Biol Rev. 2016. PMID: 27357278 Free PMC article.
-
Blocking interaction of viral gp120 and CD4-expressing T cells by single-stranded DNA aptamers.Int J Biochem Cell Biol. 2014 Jun;51:10-8. doi: 10.1016/j.biocel.2014.03.008. Epub 2014 Mar 22. Int J Biochem Cell Biol. 2014. PMID: 24661998 Free PMC article.
-
Resistance profile of the HIV-1 maturation inhibitor GSK3532795 in vitro and in a clinical study.PLoS One. 2019 Oct 17;14(10):e0224076. doi: 10.1371/journal.pone.0224076. eCollection 2019. PLoS One. 2019. PMID: 31622432 Free PMC article. Clinical Trial.
References
-
- Detels R, Munoz A, McFarlane G, Kingsley LA, Margolick JB, Giorgi J, Schrager LK, Phair JP. 1998. Effectiveness of potent antiretroviral therapy on time to AIDS and death in men with known HIV infection duration. JAMA 280:1497–1503 - PubMed
-
- Murphy EL, Collier AC, Kalish LA, Assmann SF, Para MF, Flanigan TP, Kumar PN, Mintz L, Wallach FR, Nemo GJ. 2001. Highly active antiretroviral therapy decreases mortality and morbidity in patients with advanced HIV disease. Ann. Intern. Med. 135:17–26 - PubMed
-
- Palella FJ, Jr, Delaney KM, Moorman AC, Loveless MO, Fuhrer J, Satten GA, Aschman DJ, Holmberg SD. 1998. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N. Engl. J. Med. 338:853–860 - PubMed
-
- Fernandez-Montero JV, Barreiro P, Soriano V. 2009. HIV protease inhibitors: recent clinical trials and recommendations on use. Expert Opin. Pharmacother 10:1615–1629 - PubMed
-
- Wensing AM, van Maarseveen NM, Nijhuis M. 2010. Fifteen years of HIV protease inhibitors: raising the barrier to resistance. Antivir. Res. 85:59–74 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

