Positive selection within a diatom species acts on putative protein interactions and transcriptional regulation
- PMID: 23097498
- PMCID: PMC3548312
- DOI: 10.1093/molbev/mss242
Positive selection within a diatom species acts on putative protein interactions and transcriptional regulation
Abstract
Diatoms are the most species-rich group of microalgae, and their contribution to marine primary production is important on a global scale. Diatoms can form dense blooms through rapid asexual reproduction; mutations acquired and propagated during blooms likely provide the genetic, and thus phenotypic, variability upon which natural selection may act. Positive selection was tested using genome and transcriptome-wide pair-wise comparisons of homologs in three genera of diatoms (Pseudo-nitzschia, Ditylum, and Thalassiosira) that represent decreasing phylogenetic distances. The signal of positive selection was greatest between two strains of Thalassiosira pseudonana. Further testing among seven strains of T. pseudonana yielded 809 candidate genes of positive selection, which are 7% of the protein-coding genes. Orphan genes and genes encoding protein-binding domains and transcriptional regulators were enriched within the set of positively selected genes relative to the genome as a whole. Positively selected genes were linked to the potential selective pressures of nutrient limitation and sea surface temperature based on analysis of gene expression profiles and identification of positively selected genes in subsets of strains from locations with similar environmental conditions. The identification of positively selected genes presents an opportunity to test new hypotheses in natural populations and the laboratory that integrate selected genotypes in T. pseudonana with their associated phenotypes and selective forces.
Figures
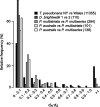
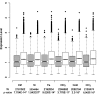


Similar articles
-
Morphological and transcriptomic evidence for ammonium induction of sexual reproduction in Thalassiosira pseudonana and other centric diatoms.PLoS One. 2017 Jul 7;12(7):e0181098. doi: 10.1371/journal.pone.0181098. eCollection 2017. PLoS One. 2017. PMID: 28686696 Free PMC article.
-
A role for the cell-wall protein silacidin in cell size of the diatom Thalassiosira pseudonana.ISME J. 2017 Nov;11(11):2452-2464. doi: 10.1038/ismej.2017.100. Epub 2017 Jul 21. ISME J. 2017. PMID: 28731468 Free PMC article.
-
The model marine diatom Thalassiosira pseudonana likely descended from a freshwater ancestor in the genus Cyclotella.BMC Evol Biol. 2011 May 14;11:125. doi: 10.1186/1471-2148-11-125. BMC Evol Biol. 2011. PMID: 21569560 Free PMC article.
-
Pseudo-nitzschia, Nitzschia, and domoic acid: New research since 2011.Harmful Algae. 2018 Nov;79:3-43. doi: 10.1016/j.hal.2018.06.001. Epub 2018 Oct 27. Harmful Algae. 2018. PMID: 30420013 Review.
-
Phylogenetic aspects of the sulfate assimilation genes from Thalassiosira pseudonana.Amino Acids. 2013 May;44(5):1253-65. doi: 10.1007/s00726-013-1462-8. Epub 2013 Jan 26. Amino Acids. 2013. PMID: 23354278 Review.
Cited by
-
Genetic characterization, structural analysis, and detection of positive selection in small heat shock proteins of Cypriniformes and Clupeiformes.Fish Physiol Biochem. 2024 Jun;50(3):843-864. doi: 10.1007/s10695-024-01337-2. Epub 2024 Apr 8. Fish Physiol Biochem. 2024. PMID: 38587724
-
Expansion Mechanisms and Evolutionary History on Genes Encoding DNA Glycosylases and Their Involvement in Stress and Hormone Signaling.Genome Biol Evol. 2016 Apr 25;8(4):1165-84. doi: 10.1093/gbe/evw067. Genome Biol Evol. 2016. PMID: 27026054 Free PMC article.
-
Dynamic responses to silicon in Thalasiossira pseudonana - Identification, characterisation and classification of signature genes and their corresponding protein motifs.Sci Rep. 2017 Jul 7;7(1):4865. doi: 10.1038/s41598-017-04921-0. Sci Rep. 2017. PMID: 28687794 Free PMC article.
-
Genome-wide identification and characterization of heat shock protein family 70 provides insight into its divergent functions on immune response and development of Paralichthys olivaceus.PeerJ. 2019 Nov 11;7:e7781. doi: 10.7717/peerj.7781. eCollection 2019. PeerJ. 2019. PMID: 31737440 Free PMC article.
-
Sucrose metabolism gene families and their biological functions.Sci Rep. 2015 Nov 30;5:17583. doi: 10.1038/srep17583. Sci Rep. 2015. PMID: 26616172 Free PMC article.
References
-
- Alba MM, Castresana J. Inverse relationship between evolutionary rate and age of mammalian genes. Mol Biol Evol. 2005;22:598–606. - PubMed
-
- Alverson AJ. Strong purifying selection in the silicon transporters of marine and freshwater diatoms. Limnol Oceanogr. 2007;52:1420–1429.
-
- Armbrust EV, Berges JA, Bowler C, et al. (45 co-authors) The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science. 2004;306:79–86. - PubMed
-
- Austin JA, Barth JA. Variation in the position of the upwelling front on the Oregon shelf. J Geophys Res. 2002;107(C11):3180.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

