Comparison of the functions of glutathionylspermidine synthetase/amidase from E. coli and its predicted homologues YgiC and YjfC
- PMID: 23097746
- PMCID: PMC3476792
Comparison of the functions of glutathionylspermidine synthetase/amidase from E. coli and its predicted homologues YgiC and YjfC
Abstract
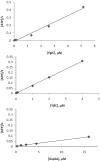
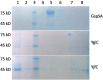
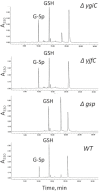
Protein function prediction is very important in establishing the roles of various proteins in bacteria; however, some proteins in the E. coli genome have their function assigned based on low percent sequence homology that does not provide reliable assignments. We have made an attempt to verify the prediction that E. coli genes ygiC and yjfC encode proteins with the same function as glutathionylspermidine synthetase/amidase (GspSA). GspSA is a bifunctional enzyme that catalyzes the ATP-dependent formation and hydrolysis of glutathionylspermidine (G-Sp), a conjugate of glutathione (GSH) and spermidine. YgiC and YjfC proteins show 51% identity between themselves and 28% identity to the synthetase domain of the GspSA enzyme. YgiC and YjfC proteins were expressed and purified, and the properties of GspSA, YgiC, and YjfC were compared. In contrast to GspSA, proteins YgiC and YjfC did not bind to G-Sp immobilized on the affinity matrix. We demonstrated that all three proteins (GspSA, YgiC and YjfC) catalyze the hydrolysis of ATP; however, YgiC and YjfC cannot synthesize G-Sp, GSH, or GSH intermediates. gsp, ygiC, and yjfC genes were eliminated from the E. coli genome to test the ability of mutant strains to synthesize G-Sp conjugate. E. coli cells deficient in GspSA do not produce G-Sp while synthesis of the conjugate is not affected in ΔygiC and ΔyjfC mutants. All together our results indicate that YgiC and YjfC are not glutathionylspermidine synthetases as predicted from the amino acid sequence analysis.
Keywords: ATP-grasp domain; ATPase; Glutathione; glutathionylspermidine; glutathionylspermidine synthetase/amidase.
Figures


Similar articles
-
Escherichia coli YgiC and YjfC Possess Peptide─Spermidine Ligase Activity.Biochemistry. 2023 Feb 21;62(4):899-911. doi: 10.1021/acs.biochem.2c00592. Epub 2023 Feb 6. Biochemistry. 2023. PMID: 36745518
-
Protein S-thiolation by Glutathionylspermidine (Gsp): the role of Escherichia coli Gsp synthetASE/amidase in redox regulation.J Biol Chem. 2010 Aug 13;285(33):25345-53. doi: 10.1074/jbc.M110.133363. Epub 2010 Jun 8. J Biol Chem. 2010. PMID: 20530482 Free PMC article.
-
Glutathionylspermidine metabolism in Escherichia coli. Purification, cloning, overproduction, and characterization of a bifunctional glutathionylspermidine synthetase/amidase.J Biol Chem. 1995 Jun 9;270(23):14031-41. doi: 10.1074/jbc.270.23.14031. J Biol Chem. 1995. PMID: 7775463
-
The CHAP domain: a large family of amidases including GSP amidase and peptidoglycan hydrolases.Trends Biochem Sci. 2003 May;28(5):234-7. doi: 10.1016/S0968-0004(03)00061-6. Trends Biochem Sci. 2003. PMID: 12765834 Review.
-
Metabolism and functions of trypanothione in the Kinetoplastida.Annu Rev Microbiol. 1992;46:695-729. doi: 10.1146/annurev.mi.46.100192.003403. Annu Rev Microbiol. 1992. PMID: 1444271 Review.
Cited by
-
A historical perspective on the multifunctional outer membrane channel protein TolC in Escherichia coli.NPJ Antimicrob Resist. 2025 Jan 25;3(1):6. doi: 10.1038/s44259-025-00078-3. NPJ Antimicrob Resist. 2025. PMID: 39863731 Free PMC article. Review.
-
Insyght: navigating amongst abundant homologues, syntenies and gene functional annotations in bacteria, it's that symbol!Nucleic Acids Res. 2014 Dec 1;42(21):e162. doi: 10.1093/nar/gku867. Epub 2014 Sep 23. Nucleic Acids Res. 2014. PMID: 25249626 Free PMC article.
References
-
- Karp PD, Keseler IM, Shearer A, Latendresse M, Krummenacker M, Paley SM, Paulsen I, Collado-Vides J, Gama-Castro S, Peralta-Gil M, Santos-Zavaleta A, Peñaloza-Spínola MI, Bonavides-Martinez C, Ingraham J. Multidimensional annotation of the Escherichia coli K-12 genome. Nucleic Acids Res. 2007;35:7577–90. - PMC - PubMed
-
- Keseler IM, Collado-Vides J, Santos-Zavaleta A, Peralta-Gil M, Gama-Castro S, Muñiz-Rascado L, Bonavides-Martinez C, Paley S, Krummenacker M, Altman T, Kaipa P, Spaulding A, Pacheco J, Latendresse M, Fulcher C, Sarker M, Shearer AG, Mackie A, Paulsen I, Gunsalus RP, Karp PD. EcoCyc: a comprehensive database of Escherichia coli biology . Nucleic Acids Res. 2011;39:D583–90. - PMC - PubMed
-
- Bollinger JM Jr, Kwon DS, Huisman GW, Kolter R, Walsh CT. Glutathionylspermidine metabo lism in Escherichia coli. Purification, cloning, overproduction, and characterization of a bifunctional glutathionylspermidine synthetase/amidase. J Biol Chem. 1995;270:14031–41. - PubMed
-
- Kwon DS, Lin CH, Chen S, Coward JK, Walsh CT, Bollinger JM Jr. Dissection of glutathionylspermidine synthetase/amidase from Escherichia coli into autonomously folding and functional synthetase and amidase domains. J Biol Chem. 1997;272:2429–36. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
