Novel epitopes identified by anti-PrP monoclonal antibodies produced following immunization of Prnp0/0 Balb/cJ mice with purified scrapie prions
- PMID: 23098297
- PMCID: PMC3482378
- DOI: 10.1089/hyb.2012.0022
Novel epitopes identified by anti-PrP monoclonal antibodies produced following immunization of Prnp0/0 Balb/cJ mice with purified scrapie prions
Abstract
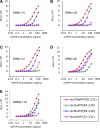
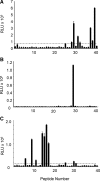
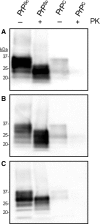
Prions, or infectious proteins, cause a class of uniformly fatal neurodegenerative diseases. Prions are composed solely of an aberrantly folded isoform (PrP(Sc)) of a normal cellular protein (PrP(C)). Shared sequence identity of PrP(Sc) with PrP(C) has limited the detection sensitivity of immunochemical assays, as antibodies specific for the disease-causing PrP(Sc) isoform have not been developed. Here we report the generation of three new monoclonal antibodies (MAbs) to PrP, which were isolated following immunization of Prnp(0/0) Balb/cJ mice with highly purified PrP(Sc) isolated from brain lipid rafts. Epitope mapping using synthetic PrP peptides revealed that the three MAbs bind different epitopes of PrP. The DRM1-31 MAb has a conformational epitope at the proposed binding site for the putative prion conversion co-factor "protein X." The DRM1-60 MAb binds a single linear epitope localized to the β2-α2 loop region of PrP, whereas DRM2-118 binds an epitope that includes sequences within the octarepeat region and near the site of N-terminal truncation of PrP(Sc) by proteinase K. Our novel anti-PrP MAbs with defined PrP epitopes may be useful in deciphering the conformational conversion of PrP(C) into PrP(Sc).
Figures





References
-
- DeArmond SJ. Mobley WC. DeMott DL. Barry RA. Beckstead JH. Prusiner SB. Changes in the localization of brain prion proteins during scrapie infection. Neurology. 1987;37:1271–1280. - PubMed
-
- Prusiner SB. Scrapie prions. Annu Rev Microbiol. 1989;43:345–374. - PubMed
-
- Caughey BW. Dong A. Bhat KS. Ernst D. Hayes SF. Caughey WS. Secondary structure analysis of the scrapie-associated protein PrP 27–30 in water by infrared spectroscopy. Biochemistry. 1991;30:7672–7680. - PubMed
-
- Prusiner SB. Scott M. Foster D. Pank M. Groth D. Mirenda C. Torchia M. Yang SL. Carlson GA. Hoppe PC. Westaway D. DeArmond SJ. Transgenic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell. 1990;63:673–686. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

