Site-specific phosphorylation of the p65 protein subunit mediates selective gene expression by differential NF-κB and RNA polymerase II promoter recruitment
- PMID: 23100252
- PMCID: PMC3537023
- DOI: 10.1074/jbc.M112.385625
Site-specific phosphorylation of the p65 protein subunit mediates selective gene expression by differential NF-κB and RNA polymerase II promoter recruitment
Abstract
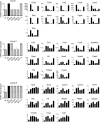
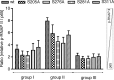
Phosphorylation of NF-κB plays an important role in modulating transcriptional activity of NF-κB independently of inhibitor of κB (IκB) proteins. For the p65 subunit, multiple phosphorylation sites have been mapped in and adjacent to both the N-terminal Rel homology domain and the C-terminal transactivation domain. Their impact on NF-κB-dependent transcription, however, has never been assessed at a broader level. In this study, we evaluate the importance of differential p65 phosphorylation on four serine acceptor sites in the Rel homology domain for the expression of an array of NF-κB-dependent genes in endothelial cells. We find that inhibition of p65 phosphorylation on these serine residues targets NF-κB activity to distinctive gene subsets in a κB enhancer element-specific context. We show that the phosphorylation-dependent alterations in gene and protein expression are reflective of the amount of p65 and phosphorylated RNA polymerase II (p-RNAP II) bound to respective gene promoter regions. Depending on the gene subset, impaired gene expression was either a result of decreased p65 promoter recruitment or of a failure of bound p65 to recruit p-RNAP II. In conclusion, our findings demonstrate that site-specific p65 phosphorylation targets NF-κB activity to particular gene subsets on a global level by influencing p65 and p-RNAP II promoter recruitment.
Figures




References
-
- Barkett M., Gilmore T. D. (1999) Control of apoptosis by Rel/NF-κB transcription factors. Oncogene 18, 6910–6924 - PubMed
-
- Hayden M. S., West A. P., Ghosh S. (2006) NF-κB and the immune response. Oncogene 25, 6758–6780 - PubMed
-
- Karin M., Lin A. (2002) NF-κB at the crossroads of life and death. Nat. Immunol. 3, 221–227 - PubMed
-
- Baeuerle P. A., Baltimore D. (1988) IκB. A specific inhibitor of the NF-κB transcription factor. Science 242, 540–546 - PubMed
-
- Henkel T., Machleidt T., Alkalay I., Krönke M., Ben-Neriah Y., Baeuerle P. A. (1993) Rapid proteolysis of IκB-α is necessary for activation of transcription factor NF-κB. Nature 365, 182–185 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

