Myocardial gene expression profiles and cardiodepressant autoantibodies predict response of patients with dilated cardiomyopathy to immunoadsorption therapy
- PMID: 23100283
- PMCID: PMC3584995
- DOI: 10.1093/eurheartj/ehs330
Myocardial gene expression profiles and cardiodepressant autoantibodies predict response of patients with dilated cardiomyopathy to immunoadsorption therapy
Abstract
Aims: Immunoadsorption with subsequent immunoglobulin G substitution (IA/IgG) represents a novel therapeutic approach in the treatment of dilated cardiomyopathy (DCM) which leads to the improvement of left ventricular ejection fraction (LVEF). However, response to this therapeutic intervention shows wide inter-individual variability. In this pilot study, we tested the value of clinical, biochemical, and molecular parameters for the prediction of the response of patients with DCM to IA/IgG.
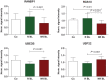
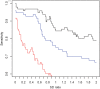
Methods and results: Forty DCM patients underwent endomyocardial biopsies (EMBs) before IA/IgG. In eight patients with normal LVEF (controls), EMBs were obtained for clinical reasons. Clinical parameters, negative inotropic activity (NIA) of antibodies on isolated rat cardiomyocytes, and gene expression profiles of EMBs were analysed. Dilated cardiomyopathy patients displaying improvement of LVEF (≥20 relative and ≥5% absolute) 6 months after IA/IgG were considered responders. Compared with non-responders (n = 16), responders (n = 24) displayed shorter disease duration (P = 0.006), smaller LV internal diameter in diastole (P = 0.019), and stronger NIA of antibodies. Antibodies obtained from controls were devoid of NIA. Myocardial gene expression patterns were different in responders and non-responders for genes of oxidative phosphorylation, mitochondrial dysfunction, hypertrophy, and ubiquitin-proteasome pathway. The integration of scores of NIA and expression levels of four genes allowed robust discrimination of responders from non-responders at baseline (BL) [sensitivity of 100% (95% CI 85.8-100%); specificity up to 100% (95% CI 79.4-100%); cut-off value: -0.28] and was superior to scores derived from antibodies, gene expression, or clinical parameters only.
Conclusion: Combined assessment of NIA of antibodies and gene expression patterns of DCM patients at BL predicts response to IA/IgG therapy and may enable appropriate selection of patients who benefit from this therapeutic intervention.
Figures



Comment in
-
New technologies, new therapies: toward personalized medicine in heart failure patients?Eur Heart J. 2013 Mar;34(9):636-7. doi: 10.1093/eurheartj/ehs432. Epub 2012 Dec 20. Eur Heart J. 2013. PMID: 23257952 No abstract available.
References
-
- Maron BJ, Towbin JA, Thiene G, Antzelevitch C, Corrado D, Arnett D, Moss AJ, Seidman CE, Young JB. Contemporary definitions and classification of the cardiomyopathies: an American Heart Association Scientific Statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; and Council on Epidemiology and Prevention. Circulation. 2006;113:1807–1816. - PubMed
-
- Kühl U, Schultheiss HP. Viral myocarditis: diagnosis, aetiology and management. Drugs. 2009;69:1287–1302. - PubMed
-
- Heymans S, Hirsch E, Anker SD, Aukrust P, Balligand JL, Cohen-Tervaert JW, Drexler H, Filippatos G, Felix SB, Gullestad L, Hilfiker-Kleiner D, Janssens S, Latini R, Neubauer G, Paulus WJ, Pieske B, Ponikowski P, Schroen B, Schultheiss HP, Tschope C, Van Bilsen M, Zannad F, McMurray J, Shah AM. Inflammation as a therapeutic target in heart failure? A scientific statement from the Translational Research Committee of the Heart Failure Association of the European Society of Cardiology. Eur J Heart Fail. 2009;11:119–129. - PMC - PubMed
-
- Cihakova D, Rose NR. Pathogenesis of myocarditis and dilated cardiomyopathy. Adv Immunol. 2008;99:95–114. - PubMed
-
- Kallwellis-Opara A, Dorner A, Poller WC, Noutsias M, Kuhl U, Schultheiss HP, Pauschinger M. Autoimmunological features in inflammatory cardiomyopathy. Clin Res Cardiol. 2007;96:469–480. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

