RAB-5 and RAB-10 cooperate to regulate neuropeptide release in Caenorhabditis elegans
- PMID: 23100538
- PMCID: PMC3503202
- DOI: 10.1073/pnas.1203306109
RAB-5 and RAB-10 cooperate to regulate neuropeptide release in Caenorhabditis elegans
Abstract
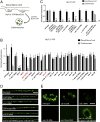
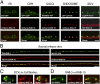
Neurons secrete neuropeptides from dense core vesicles (DCVs) to modulate neuronal activity. Little is known about how neurons manage to differentially regulate the release of synaptic vesicles (SVs) and DCVs. To analyze this, we screened all Caenorhabditis elegans Rab GTPases and Tre2/Bub2/Cdc16 (TBC) domain containing GTPase-activating proteins (GAPs) for defects in DCV release from C. elegans motoneurons. rab-5 and rab-10 mutants show severe defects in DCV secretion, whereas SV exocytosis is unaffected. We identified TBC-2 and TBC-4 as putative GAPs for RAB-5 and RAB-10, respectively. Multiple Rabs and RabGAPs are typically organized in cascades that confer directionality to membrane-trafficking processes. We show here that the formation of release-competent DCVs requires a reciprocal exclusion cascade coupling RAB-5 and RAB-10, in which each of the two Rabs recruits the other's GAP molecule. This contributes to a separation of RAB-5 and RAB-10 domains at the Golgi-endosomal interface, which is lost when either of the two GAPs is inactivated. Taken together, our data suggest that RAB-5 and RAB-10 cooperate to locally exclude each other at an essential stage during DCV sorting.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
TBC-8, a putative RAB-2 GAP, regulates dense core vesicle maturation in Caenorhabditis elegans.PLoS Genet. 2012;8(5):e1002722. doi: 10.1371/journal.pgen.1002722. Epub 2012 May 24. PLoS Genet. 2012. PMID: 22654674 Free PMC article.
-
UNC-108/RAB-2 and its effector RIC-19 are involved in dense core vesicle maturation in Caenorhabditis elegans.J Cell Biol. 2009 Sep 21;186(6):897-914. doi: 10.1083/jcb.200902096. J Cell Biol. 2009. PMID: 19797081 Free PMC article.
-
A novel CaM kinase II pathway controls the location of neuropeptide release from Caenorhabditis elegans motor neurons.Genetics. 2014 Mar;196(3):745-65. doi: 10.1534/genetics.113.158568. Genetics. 2014. PMID: 24653209 Free PMC article.
-
Regulation of secretory vesicle traffic by Rab small GTPases.Cell Mol Life Sci. 2008 Sep;65(18):2801-13. doi: 10.1007/s00018-008-8351-4. Cell Mol Life Sci. 2008. PMID: 18726178 Free PMC article. Review.
-
Neurotransmitter release mechanisms studied in Caenorhabditis elegans.Cell Calcium. 2012 Sep-Oct;52(3-4):289-95. doi: 10.1016/j.ceca.2012.03.005. Epub 2012 Apr 20. Cell Calcium. 2012. PMID: 22521667 Review.
Cited by
-
Luqin-like RYamide peptides regulate food-evoked responses in C. elegans.Elife. 2017 Aug 29;6:e28877. doi: 10.7554/eLife.28877. Elife. 2017. PMID: 28847365 Free PMC article.
-
Crosstalk between the Rho and Rab family of small GTPases in neurodegenerative disorders.Front Cell Neurosci. 2023 Jan 27;17:1084769. doi: 10.3389/fncel.2023.1084769. eCollection 2023. Front Cell Neurosci. 2023. PMID: 36779014 Free PMC article. Review.
-
Chronic treatment with fluoride affects the jejunum: insights from proteomics and enteric innervation analysis.Sci Rep. 2018 Feb 16;8(1):3180. doi: 10.1038/s41598-018-21533-4. Sci Rep. 2018. PMID: 29453425 Free PMC article.
-
Large-scale transcriptome changes in the process of long-term visual memory formation in the bumblebee, Bombus terrestris.Sci Rep. 2018 Jan 11;8(1):534. doi: 10.1038/s41598-017-18836-3. Sci Rep. 2018. PMID: 29323174 Free PMC article.
-
Axonal Endoplasmic Reticulum Dynamics and Its Roles in Neurodegeneration.Front Neurosci. 2020 Jan 29;14:48. doi: 10.3389/fnins.2020.00048. eCollection 2020. Front Neurosci. 2020. PMID: 32116502 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

