Plant plasma membrane-bound staphylococcal-like DNases as a novel class of eukaryotic nucleases
- PMID: 23102437
- PMCID: PMC3505149
- DOI: 10.1186/1471-2229-12-195
Plant plasma membrane-bound staphylococcal-like DNases as a novel class of eukaryotic nucleases
Abstract
Background: The activity of degradative nucleases responsible for genomic DNA digestion has been observed in all kingdoms of life. It is believed that the main function of DNA degradation occurring during plant programmed cell death is redistribution of nucleic acid derived products such as nitrogen, phosphorus and nucleotide bases. Plant degradative nucleases that have been studied so far belong mainly to the S1-type family and were identified in cellular compartments containing nucleic acids or in the organelles where they are stored before final application. However, the explanation of how degraded DNA components are exported from the dying cells for further reutilization remains open.
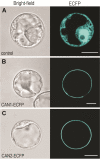
Results: Bioinformatic and experimental data presented in this paper indicate that two Arabidopsis staphylococcal-like nucleases, named CAN1 and CAN2, are anchored to the cell membrane via N-terminal myristoylation and palmitoylation modifications. Both proteins possess a unique hybrid structure in their catalytic domain consisting of staphylococcal nuclease-like and tRNA synthetase anticodon binding-like motifs. They are neutral, Ca2+-dependent nucleaces showing a different specificity toward the ssDNA, dsDNA and RNA substrates. A study of microarray experiments and endogenous nuclease activity revealed that expression of CAN1 gene correlates with different forms of programmed cell death, while the CAN2 gene is constitutively expressed.
Conclusions: In this paper we present evidence showing that two plant staphylococcal-like nucleases belong to a new, as yet unidentified class of eukaryotic nucleases, characterized by unique plasma membrane localization. The identification of this class of nucleases indicates that plant cells possess additional, so far uncharacterized, mechanisms responsible for DNA and RNA degradation. The potential functions of these nucleases in relation to their unique intracellular location are discussed.
Figures






References
-
- Guo Y, Cai Z, Gan S. Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ. 2004;27:521–549. doi: 10.1111/j.1365-3040.2003.01158.x. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

