Direct imaging of RecA nucleation and growth on single molecules of SSB-coated ssDNA
- PMID: 23103864
- PMCID: PMC4112059
- DOI: 10.1038/nature11598
Direct imaging of RecA nucleation and growth on single molecules of SSB-coated ssDNA
Abstract
Escherichia coli RecA is the defining member of a ubiquitous class of DNA strand-exchange proteins that are essential for homologous recombination, a pathway that maintains genomic integrity by repairing broken DNA. To function, filaments of RecA must nucleate and grow on single-stranded DNA (ssDNA) in direct competition with ssDNA-binding protein (SSB), which rapidly binds and continuously sequesters ssDNA, kinetically blocking RecA assembly. This dynamic self-assembly on a DNA lattice, in competition with another protein, is unique for the RecA family compared to other filament-forming proteins such as actin and tubulin. The complexity of this process has hindered our understanding of RecA filament assembly because ensemble measurements cannot reliably distinguish between the nucleation and growth phases, despite extensive and diverse attempts. Previous single-molecule assays have measured the nucleation and growth of RecA--and its eukaryotic homologue RAD51--on naked double-stranded DNA and ssDNA; however, the template for RecA self-assembly in vivo is SSB-coated ssDNA. Using single-molecule microscopy, here we directly visualize RecA filament assembly on single molecules of SSB-coated ssDNA, simultaneously measuring nucleation and growth. We establish that a dimer of RecA is required for nucleation, followed by growth of the filament through monomer addition, consistent with the finding that nucleation, but not growth, is modulated by nucleotide and magnesium ion cofactors. Filament growth is bidirectional, albeit faster in the 5'→3' direction. Both nucleation and growth are repressed at physiological conditions, highlighting the essential role of recombination mediators in potentiating assembly in vivo. We define a two-step kinetic mechanism in which RecA nucleates on transiently exposed ssDNA during SSB sliding and/or partial dissociation (DNA unwrapping) and then the RecA filament grows. We further demonstrate that the recombination mediator protein pair, RecOR (RecO and RecR), accelerates both RecA nucleation and filament growth, and that the introduction of RecF further stimulates RecA nucleation.
Conflict of interest statement
Figures

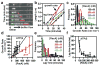
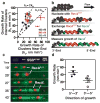

Comment in
-
Biochemistry: A glimpse of molecular competition.Nature. 2012 Nov 8;491(7423):198-200. doi: 10.1038/nature11639. Epub 2012 Oct 24. Nature. 2012. PMID: 23103870 No abstract available.
References
-
- Kowalczykowski SC, Eggleston AK. Homologous pairing and DNA strand-exchange proteins. Annu Rev Biochem. 1994;63:991–1043. - PubMed
-
- Kowalczykowski SC. Biochemistry of genetic recombination: Energetics and mechanism of DNA strand exchange. Annu Rev Biophys Biophys Chem. 1991;20:539–575. - PubMed
-
- Kowalczykowski SC, Clow J, Somani R, Varghese A. Effects of the Escherichia coli SSB protein on the binding of Escherichia coli RecA protein to single-stranded DNA: Demonstration of competitive binding and the lack of a specific protein-protein interaction. J Mol Biol. 1987;193:81–95. - PubMed
-
- Thresher RJ, Christiansen G, Griffith JD. Assembly of presynaptic filaments. Factors affecting the assembly of RecA protein onto single-stranded DNA. J Mol Biol. 1988;201:101–113. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

