Inhibition of RNA lariat debranching enzyme suppresses TDP-43 toxicity in ALS disease models
- PMID: 23104007
- PMCID: PMC3510335
- DOI: 10.1038/ng.2434
Inhibition of RNA lariat debranching enzyme suppresses TDP-43 toxicity in ALS disease models
Abstract
Amyotrophic lateral sclerosis (ALS) is a devastating neurodegenerative disease primarily affecting motor neurons. Mutations in the gene encoding TDP-43 cause some forms of the disease, and cytoplasmic TDP-43 aggregates accumulate in degenerating neurons of most individuals with ALS. Thus, strategies aimed at targeting the toxicity of cytoplasmic TDP-43 aggregates may be effective. Here, we report results from two genome-wide loss-of-function TDP-43 toxicity suppressor screens in yeast. The strongest suppressor of TDP-43 toxicity was deletion of DBR1, which encodes an RNA lariat debranching enzyme. We show that, in the absence of Dbr1 enzymatic activity, intronic lariats accumulate in the cytoplasm and likely act as decoys to sequester TDP-43, preventing it from interfering with essential cellular RNAs and RNA-binding proteins. Knockdown of Dbr1 in a human neuronal cell line or in primary rat neurons is also sufficient to rescue TDP-43 toxicity. Our findings provide insight into TDP-43-mediated cytotoxicity and suggest that decreasing Dbr1 activity could be a potential therapeutic approach for ALS.
Conflict of interest statement
A.D.G. is an inventor on patents and patent applications that have been licensed to FoldRx.
Figures




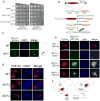
References
-
- Boillee S, Vande Velde C, Cleveland DW. ALS: a disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52:39–59. - PubMed
-
- Rosen D, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. - PubMed
-
- Bruijn LI, et al. Aggregation and motor neuron toxicity of an ALS-linked SOD1 mutant independent from wild-type SOD1. Science. 1998;281:1851–4. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DP2 OD004417/OD/NIH HHS/United States
- NS072233/NS/NINDS NIH HHS/United States
- GM081879/GM/NIGMS NIH HHS/United States
- GM084279/GM/NIGMS NIH HHS/United States
- R01 NS039074/NS/NINDS NIH HHS/United States
- P50 GM081879/GM/NIGMS NIH HHS/United States
- 1DP2OD004417/OD/NIH HHS/United States
- NS045491/NS/NINDS NIH HHS/United States
- K08 NS072233/NS/NINDS NIH HHS/United States
- R01 GM098101/GM/NIGMS NIH HHS/United States
- GM098101/GM/NIGMS NIH HHS/United States
- DP2 OD002177/OD/NIH HHS/United States
- R01 NS065317/NS/NINDS NIH HHS/United States
- 1DP2OD002177/OD/NIH HHS/United States
- NS39074/NS/NINDS NIH HHS/United States
- 2P01AG02074/AG/NIA NIH HHS/United States
- RR18928/RR/NCRR NIH HHS/United States
- GM084448/GM/NIGMS NIH HHS/United States
- P30 AI027763/AI/NIAID NIH HHS/United States
- R01 GM084448/GM/NIGMS NIH HHS/United States
- R21 NS067354/NS/NINDS NIH HHS/United States
- R01 GM084279/GM/NIGMS NIH HHS/United States
- NS065317/NS/NINDS NIH HHS/United States
- C06 RR018928/RR/NCRR NIH HHS/United States
- NS067354/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

