Nonlinear fitness consequences of variation in expression level of a eukaryotic gene
- PMID: 23104081
- PMCID: PMC3548308
- DOI: 10.1093/molbev/mss248
Nonlinear fitness consequences of variation in expression level of a eukaryotic gene
Abstract
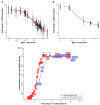
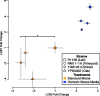
Levels of gene expression show considerable variation in eukaryotes, but no fine-scale maps have been made of the fitness consequences of such variation in controlled genetic backgrounds and environments. To address this, we assayed fitness at many levels of up- and down-regulated expression of a single essential gene, LCB2, involved in sphingolipid synthesis in budding yeast Saccharomyces cerevisiae. Reduced LCB2 expression rapidly decreases cellular fitness, yet increased expression has little effect. The wild-type expression level is therefore perched on the edge of a nonlinear fitness cliff. LCB2 is upregulated when cells are exposed to osmotic stress; consistent with this, the entire fitness curve is shifted upward to higher expression under osmotic stress, illustrating the selective force behind gene regulation. Expression levels of LCB2 are lower in wild yeast strains than in the experimental lab strain, suggesting that higher levels in the lab strain may be idiosyncratic. Reports indicate that the effect sizes of alleles contributing to variation in complex phenotypes differ among environments and genetic backgrounds; our results suggest that such differences may be explained as simple shifts in the position of nonlinear fitness curves.
Figures

References
-
- Bayer TS. Using synthetic biology to understand the evolution of gene expression. Curr Biol. 2010;20:R772–R779. - PubMed
-
- Bielawski J, Szulc ZM, Hannun YA, Bielawska A. Simultaneous quantitative analysis of bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Methods. 2006;39:82–91. - PubMed
-
- Birchler JA, Bhadra U, Bhadra MP, Auger DL. Dosage-dependent gene regulation in multicellular eukaryotes: implications for dosage compensation, aneuploid syndromes, and quantitative traits. Dev Biol. 2001;234:275–288. - PubMed
-
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. - PubMed
-
- Brem RB, Yvert Gl, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

