Understanding protein aggregation from the view of monomer dynamics
- PMID: 23104145
- PMCID: PMC4834696
- DOI: 10.1039/c2mb25334h
Understanding protein aggregation from the view of monomer dynamics
Abstract
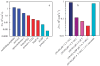
Much work in recent years has been devoted to understanding the complex process of protein aggregation. This review looks at the earliest stages of aggregation, long before the formation of fibrils that are the hallmark of many aggregation-based diseases, and proposes that the first steps are controlled by the reconfiguration dynamics of the monomer. When reconfiguration is much faster or much slower than bimolecular diffusion, then aggregation is slow, but when they are similar, aggregation is fast. The experimental evidence for this model is reviewed and the prospects for small molecule aggregation inhibitors to prevent disease are discussed.
Figures

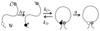
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

