Arabidopsis bZIP16 transcription factor integrates light and hormone signaling pathways to regulate early seedling development
- PMID: 23104829
- PMCID: PMC3517232
- DOI: 10.1105/tpc.112.105478
Arabidopsis bZIP16 transcription factor integrates light and hormone signaling pathways to regulate early seedling development
Abstract
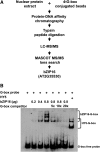
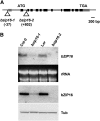
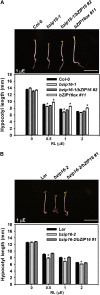
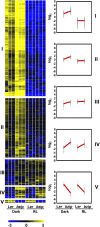
Transcriptomic adjustment plays an important role in Arabidopsis thaliana seed germination and deetiolation in response to environmental light signals. The G-box cis-element is commonly present in promoters of genes that respond positively or negatively to the light signal. In pursuing additional transcriptional regulators that modulate light-mediated transcriptome changes, we identified bZIP16, a basic region/Leu zipper motif transcription factor, by G-box DNA affinity chromatography. We confirmed that bZIP16 has G-box-specific binding activity. Analysis of bzip16 mutants revealed that bZIP16 is a negative regulator in light-mediated inhibition of cell elongation but a positive regulator in light-regulated seed germination. Transcriptome analysis supported that bZIP16 is primarily a transcriptional repressor regulating light-, gibberellic acid (GA)-, and abscisic acid (ABA)-responsive genes. Chromatin immunoprecipitation analysis revealed that bZIP16 could directly target ABA-responsive genes and RGA-like2, a DELLA gene in the GA signaling pathway. bZIP16 could also indirectly repress the expression of phytochrome interacting factoR3-like5, which encodes a basic helix-loop-helix protein coordinating hormone responses during seed germination. By repressing the expression of these genes, bZIP16 functions to promote seed germination and hypocotyl elongation during the early stages of Arabidopsis seedling development.
Figures





References
-
- Agresti A. (1992). A survey of exact inference for contingency tables. Stat. Sci. 7: 131–153
-
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 - PubMed
-
- Ang L.H., Chattopadhyay S., Wei N., Oyama T., Okada K., Batschauer A., Deng X.W. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1: 213–222 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

