Gas-phase studies of substrates for the DNA mismatch repair enzyme MutY
- PMID: 23106240
- PMCID: PMC4204490
- DOI: 10.1021/ja309082k
Gas-phase studies of substrates for the DNA mismatch repair enzyme MutY
Abstract
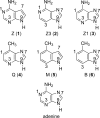
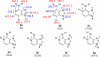
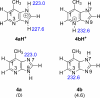
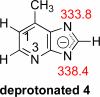
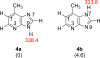
The gas-phase thermochemical properties (tautomeric energies, acidity, and proton affinity) have been measured and calculated for adenine and six adenine analogues that were designed to test features of the catalytic mechanism used by the adenine glycosylase MutY. The gas-phase intrinsic properties are correlated to possible excision mechanisms and MutY excision rates to gain insight into the MutY mechanism. The data support a mechanism involving protonation at N7 and hydrogen bonding to N3 of adenine. We also explored the acid-catalyzed (non-enzymatic) depurination of these substrates, which appears to follow a different mechanism than that employed by MutY, which we elucidate using calculations.
Figures

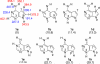

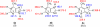
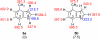






References
-
- David SS, Williams SD. Chem. Rev. 1998;98:1221–1261. - PubMed
-
- Lindahl T. Nature. 1993;362:709–715. - PubMed
-
- Klaunig JE, Kamendulis LM. Annu. Rev. Pharmacol. Toxicol. 2004;44:239–267. - PubMed
-
- Neeley WL, Essigmann JM. Chem. Res. Toxicol. 2006;19:491–505. - PubMed
-
- Shigenaga MK, Park J-W, Cundy KC, Gimeno CJ, Ames BN. Methods Enzymol. 1990;186:521–530. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

